Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6448
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7557 | 3.400e-03 | 5.480e-04 | 7.658e-01 | 1.427e-06 |
|---|
| Loi | 0.2298 | 8.108e-02 | 1.158e-02 | 4.302e-01 | 4.038e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.580e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.487e-03 | 0.000e+00 |
|---|
| Wang | 0.2609 | 2.767e-03 | 4.463e-02 | 4.001e-01 | 4.941e-05 |
|---|
Expression data for subnetwork 6448 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
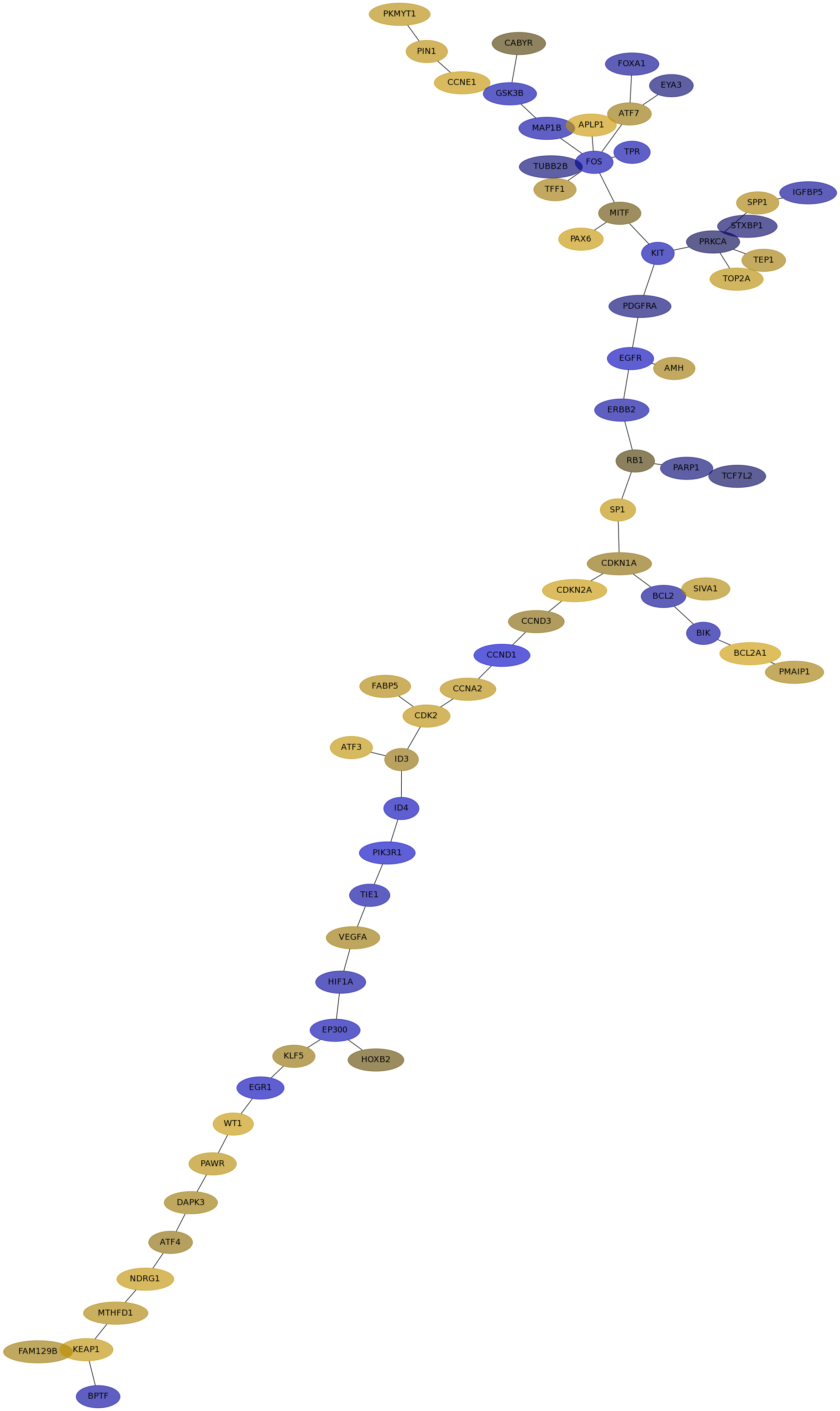
Score for each gene in subnetwork 6448 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| BPTF |   | 4 | 440 | 318 | 337 | -0.098 | 0.159 | 0.132 | undef | 0.030 |
|---|
| NDRG1 |   | 6 | 301 | 658 | 646 | 0.222 | 0.143 | -0.040 | -0.018 | 0.242 |
|---|
| FAM129B |   | 1 | 1195 | 1967 | 1973 | 0.101 | 0.243 | undef | undef | undef |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| BIK |   | 1 | 1195 | 1967 | 1973 | -0.097 | 0.133 | -0.179 | -0.092 | -0.071 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PAWR |   | 1 | 1195 | 1967 | 1973 | 0.173 | 0.224 | 0.034 | 0.124 | 0.137 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| CCNE1 |   | 4 | 440 | 1083 | 1067 | 0.245 | -0.141 | -0.077 | undef | -0.018 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| APLP1 |   | 8 | 222 | 727 | 703 | 0.284 | 0.043 | -0.105 | -0.121 | -0.190 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| PMAIP1 |   | 4 | 440 | 1055 | 1035 | 0.118 | -0.105 | 0.057 | undef | -0.047 |
|---|
| SIVA1 |   | 3 | 557 | 1055 | 1043 | 0.157 | -0.103 | 0.190 | undef | -0.042 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| CCND3 |   | 1 | 1195 | 1967 | 1973 | 0.053 | 0.056 | 0.015 | -0.123 | -0.171 |
|---|
| AMH |   | 10 | 167 | 366 | 362 | 0.103 | 0.056 | 0.113 | -0.187 | 0.301 |
|---|
| MITF |   | 5 | 360 | 696 | 680 | 0.027 | 0.032 | 0.065 | 0.145 | 0.183 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| STXBP1 |   | 5 | 360 | 1629 | 1587 | -0.027 | 0.078 | 0.180 | 0.143 | 0.043 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MTHFD1 |   | 2 | 743 | 1604 | 1588 | 0.139 | -0.266 | 0.078 | 0.066 | 0.124 |
|---|
| CABYR |   | 1 | 1195 | 1967 | 1973 | 0.015 | -0.003 | -0.046 | undef | -0.088 |
|---|
| ATF4 |   | 1 | 1195 | 1967 | 1973 | 0.065 | 0.136 | -0.041 | 0.041 | -0.032 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| TEP1 |   | 2 | 743 | 1967 | 1945 | 0.111 | 0.126 | -0.039 | 0.182 | 0.134 |
|---|
| FABP5 |   | 3 | 557 | 1534 | 1516 | 0.152 | -0.177 | 0.225 | -0.118 | 0.141 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| PKMYT1 |   | 7 | 256 | 1510 | 1463 | 0.175 | -0.143 | 0.046 | 0.105 | -0.070 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| DAPK3 |   | 1 | 1195 | 1967 | 1973 | 0.092 | 0.145 | 0.010 | undef | 0.184 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TFF1 |   | 7 | 256 | 957 | 928 | 0.106 | 0.022 | 0.088 | -0.033 | -0.244 |
|---|
| ID3 |   | 9 | 196 | 658 | 637 | 0.072 | -0.006 | 0.192 | -0.023 | 0.057 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| KEAP1 |   | 2 | 743 | 1604 | 1588 | 0.218 | 0.011 | -0.155 | -0.205 | -0.031 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| BCL2A1 |   | 4 | 440 | 764 | 761 | 0.298 | -0.120 | -0.087 | -0.199 | -0.043 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6448 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.589E-13 | 1.121E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.993E-11 | 2.435E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.025E-09 | 4.906E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.465E-08 | 1.506E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.422E-08 | 2.161E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.067E-08 | 2.47E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.323E-08 | 3.254E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.15E-07 | 3.512E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.719E-07 | 4.667E-05 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.719E-07 | 4.201E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.175E-07 | 4.831E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.463E-13 | 2.036E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.83E-11 | 3.404E-08 |
|---|
| interphase | GO:0051325 |  | 3.875E-11 | 3.108E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.691E-09 | 5.829E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.267E-08 | 1.572E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 5.86E-08 | 2.35E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.585E-08 | 2.951E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.731E-08 | 2.926E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.319E-07 | 3.525E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.277E-07 | 5.478E-05 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 2.277E-07 | 4.98E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.724E-12 | 8.565E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.052E-10 | 1.209E-07 |
|---|
| interphase | GO:0051325 |  | 1.327E-10 | 1.018E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.293E-08 | 1.318E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.407E-08 | 2.027E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.772E-08 | 3.363E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.299E-07 | 7.554E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.595E-07 | 1.034E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.078E-07 | 1.042E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.218E-07 | 1.43E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.094E-06 | 2.287E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 5.078E-09 | 9.358E-06 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.204E-08 | 1.11E-05 |
|---|
| regulation of cell motion | GO:0051270 |  | 1.204E-08 | 7.399E-06 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.478E-08 | 2.063E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 7.405E-08 | 2.73E-05 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 1.073E-07 | 3.296E-05 |
|---|
| regulation of anatomical structure morphogenesis | GO:0022603 |  | 4.973E-07 | 1.309E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 9.392E-07 | 2.164E-04 |
|---|
| lung development | GO:0030324 |  | 1.952E-06 | 3.997E-04 |
|---|
| pigmentation | GO:0043473 |  | 2.208E-06 | 4.07E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 2.556E-06 | 4.282E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.724E-12 | 8.565E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.052E-10 | 1.209E-07 |
|---|
| interphase | GO:0051325 |  | 1.327E-10 | 1.018E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.293E-08 | 1.318E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.407E-08 | 2.027E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.772E-08 | 3.363E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.299E-07 | 7.554E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.595E-07 | 1.034E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.078E-07 | 1.042E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.218E-07 | 1.43E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.094E-06 | 2.287E-04 |
|---|



