Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6439
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7557 | 3.402e-03 | 5.480e-04 | 7.659e-01 | 1.428e-06 |
|---|
| Loi | 0.2299 | 8.097e-02 | 1.155e-02 | 4.299e-01 | 4.020e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.586e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.490e-03 | 0.000e+00 |
|---|
| Wang | 0.2608 | 2.789e-03 | 4.482e-02 | 4.010e-01 | 5.013e-05 |
|---|
Expression data for subnetwork 6439 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
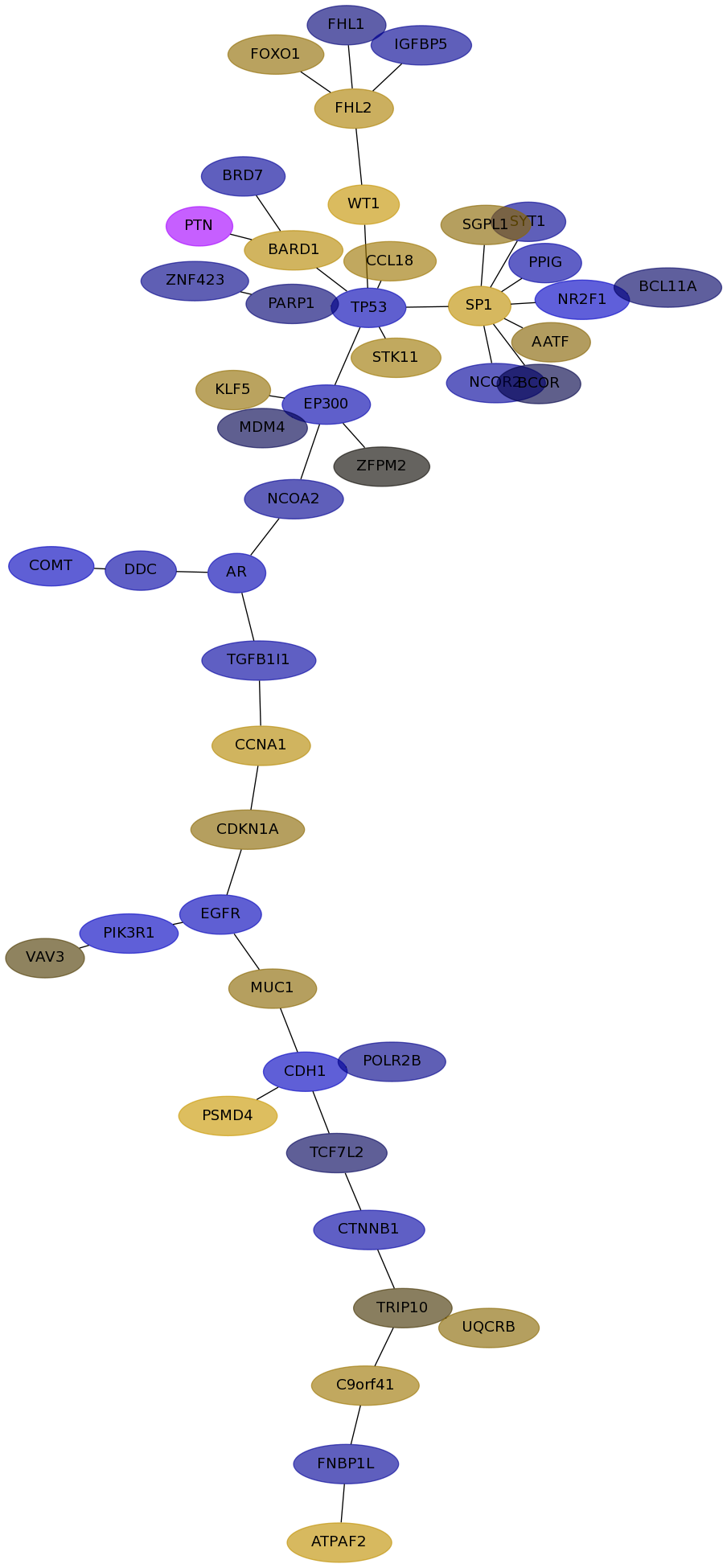
Score for each gene in subnetwork 6439 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| FOXO1 |   | 3 | 557 | 590 | 598 | 0.072 | 0.094 | -0.021 | -0.147 | 0.221 |
|---|
| TRIP10 |   | 5 | 360 | 1034 | 1004 | 0.012 | 0.154 | 0.049 | undef | 0.144 |
|---|
| UQCRB |   | 2 | 743 | 1294 | 1286 | 0.062 | -0.029 | 0.072 | 0.086 | 0.115 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| C9orf41 |   | 2 | 743 | 1294 | 1286 | 0.100 | -0.105 | undef | undef | undef |
|---|
| BCOR |   | 1 | 1195 | 2121 | 2127 | -0.013 | 0.011 | -0.067 | 0.229 | 0.053 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| SGPL1 |   | 1 | 1195 | 2121 | 2127 | 0.064 | -0.048 | -0.182 | 0.182 | -0.150 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| COMT |   | 1 | 1195 | 2121 | 2127 | -0.208 | -0.061 | 0.026 | 0.094 | -0.227 |
|---|
| ZNF423 |   | 2 | 743 | 1165 | 1163 | -0.061 | 0.100 | 0.017 | undef | 0.205 |
|---|
| POLR2B |   | 1 | 1195 | 2121 | 2127 | -0.064 | 0.012 | 0.095 | -0.018 | -0.072 |
|---|
| DDC |   | 1 | 1195 | 2121 | 2127 | -0.121 | 0.157 | -0.078 | -0.113 | 0.114 |
|---|
| NCOA2 |   | 3 | 557 | 1165 | 1151 | -0.078 | 0.080 | -0.159 | 0.104 | 0.057 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| AATF |   | 1 | 1195 | 2121 | 2127 | 0.056 | -0.163 | 0.053 | 0.092 | -0.036 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| ZFPM2 |   | 5 | 360 | 525 | 518 | 0.001 | 0.092 | 0.113 | -0.099 | 0.379 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| BCL11A |   | 2 | 743 | 1346 | 1333 | -0.026 | 0.035 | 0.064 | undef | -0.087 |
|---|
| ATPAF2 |   | 1 | 1195 | 2121 | 2127 | 0.224 | -0.040 | -0.173 | undef | 0.069 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| STK11 |   | 2 | 743 | 2053 | 2031 | 0.102 | -0.125 | -0.080 | -0.022 | -0.079 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| FNBP1L |   | 3 | 557 | 1294 | 1276 | -0.088 | 0.204 | undef | 0.154 | undef |
|---|
| CCL18 |   | 1 | 1195 | 2121 | 2127 | 0.098 | -0.189 | -0.168 | -0.170 | 0.071 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
GO Enrichment output for subnetwork 6439 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.451E-10 | 1.087E-06 |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 3.744E-09 | 4.573E-06 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.842E-07 | 2.315E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.861E-07 | 2.358E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 4.407E-07 | 2.153E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.264E-06 | 5.147E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 3.518E-06 | 1.228E-03 |
|---|
| catechol metabolic process | GO:0009712 |  | 5.517E-06 | 1.685E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 6.346E-06 | 1.722E-03 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 1.028E-05 | 2.51E-03 |
|---|
| aerobic respiration | GO:0009060 |  | 1.339E-05 | 2.973E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.004E-10 | 9.633E-07 |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 5.581E-09 | 6.714E-06 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.345E-07 | 2.682E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 3.825E-07 | 2.301E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 5.458E-07 | 2.627E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 6.05E-07 | 2.426E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.706E-06 | 5.863E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 4.745E-06 | 1.427E-03 |
|---|
| catechol metabolic process | GO:0009712 |  | 8.162E-06 | 2.182E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 9.386E-06 | 2.258E-03 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 1.384E-05 | 3.028E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 1.325E-08 | 3.046E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 8.205E-06 | 9.436E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.088E-06 | 6.968E-03 |
|---|
| catechol metabolic process | GO:0009712 |  | 1.417E-05 | 8.148E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 1.646E-05 | 7.573E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 1.721E-05 | 6.597E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 2.583E-05 | 8.486E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.221E-05 | 9.26E-03 |
|---|
| aerobic respiration | GO:0009060 |  | 3.635E-05 | 9.289E-03 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 5.766E-05 | 0.01326207 |
|---|
| ATP synthesis coupled electron transport | GO:0042773 |  | 6.983E-05 | 0.01460068 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.553E-06 | 4.704E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 1.702E-05 | 0.01568727 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 2.095E-05 | 0.01287302 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.732E-05 | 0.02180421 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.455E-04 | 0.05364358 |
|---|
| prostate gland development | GO:0030850 |  | 1.702E-04 | 0.05228811 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 1.702E-04 | 0.04481838 |
|---|
| T cell lineage commitment | GO:0002360 |  | 1.702E-04 | 0.03921608 |
|---|
| peptidyl-lysine modification | GO:0018205 |  | 1.702E-04 | 0.03485874 |
|---|
| negative regulation of neuroblast proliferation | GO:0007406 |  | 1.702E-04 | 0.03137287 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.84E-04 | 0.03083407 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 1.325E-08 | 3.046E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 8.205E-06 | 9.436E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.088E-06 | 6.968E-03 |
|---|
| catechol metabolic process | GO:0009712 |  | 1.417E-05 | 8.148E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 1.646E-05 | 7.573E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 1.721E-05 | 6.597E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 2.583E-05 | 8.486E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.221E-05 | 9.26E-03 |
|---|
| aerobic respiration | GO:0009060 |  | 3.635E-05 | 9.289E-03 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 5.766E-05 | 0.01326207 |
|---|
| ATP synthesis coupled electron transport | GO:0042773 |  | 6.983E-05 | 0.01460068 |
|---|



