Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6429
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7557 | 3.399e-03 | 5.480e-04 | 7.658e-01 | 1.426e-06 |
|---|
| Loi | 0.2298 | 8.101e-02 | 1.155e-02 | 4.300e-01 | 4.025e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.586e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.495e-03 | 0.000e+00 |
|---|
| Wang | 0.2608 | 2.787e-03 | 4.480e-02 | 4.009e-01 | 5.005e-05 |
|---|
Expression data for subnetwork 6429 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
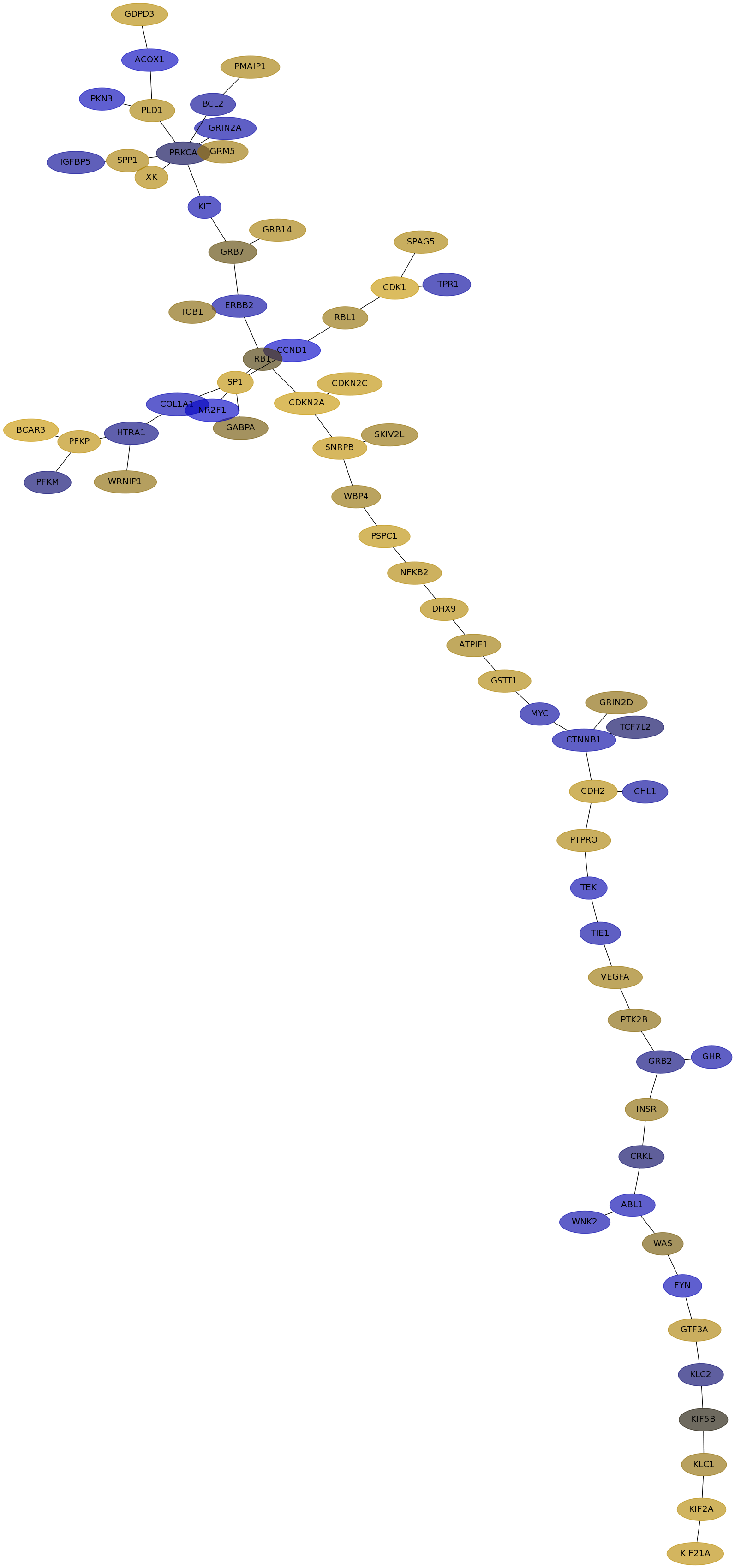
Score for each gene in subnetwork 6429 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| PTPRO |   | 2 | 743 | 1988 | 1970 | 0.133 | -0.086 | -0.058 | undef | 0.134 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| KLC2 |   | 2 | 743 | 1055 | 1059 | -0.029 | 0.017 | 0.185 | undef | 0.138 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| CRKL |   | 6 | 301 | 780 | 764 | -0.023 | 0.056 | 0.271 | -0.025 | -0.008 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PMAIP1 |   | 4 | 440 | 1055 | 1035 | 0.118 | -0.105 | 0.057 | undef | -0.047 |
|---|
| CHL1 |   | 2 | 743 | 842 | 854 | -0.088 | 0.161 | 0.202 | 0.103 | -0.105 |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| WBP4 |   | 1 | 1195 | 2111 | 2118 | 0.077 | -0.023 | 0.033 | 0.066 | -0.062 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| SKIV2L |   | 1 | 1195 | 2111 | 2118 | 0.072 | 0.031 | -0.070 | 0.049 | 0.179 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| COL1A1 |   | 3 | 557 | 552 | 558 | -0.156 | 0.244 | -0.021 | -0.081 | 0.086 |
|---|
| SNRPB |   | 2 | 743 | 478 | 490 | 0.212 | 0.123 | -0.006 | -0.136 | 0.091 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| KIF2A |   | 1 | 1195 | 2111 | 2118 | 0.173 | 0.001 | 0.052 | -0.190 | 0.023 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| WAS |   | 1 | 1195 | 2111 | 2118 | 0.035 | -0.136 | -0.075 | -0.201 | 0.053 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| PTK2B |   | 1 | 1195 | 2111 | 2118 | 0.055 | -0.169 | -0.095 | undef | -0.217 |
|---|
| PFKP |   | 6 | 301 | 83 | 105 | 0.195 | -0.076 | -0.040 | undef | 0.094 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| DHX9 |   | 1 | 1195 | 2111 | 2118 | 0.161 | 0.018 | 0.114 | 0.125 | -0.031 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| KLC1 |   | 4 | 440 | 1055 | 1035 | 0.071 | 0.189 | 0.204 | 0.139 | 0.132 |
|---|
| GTF3A |   | 2 | 743 | 1055 | 1059 | 0.138 | -0.150 | 0.224 | undef | -0.148 |
|---|
| SPAG5 |   | 1 | 1195 | 2111 | 2118 | 0.132 | -0.243 | 0.031 | 0.017 | 0.084 |
|---|
| WRNIP1 |   | 3 | 557 | 83 | 117 | 0.064 | -0.025 | 0.162 | undef | -0.115 |
|---|
| KIF5B |   | 2 | 743 | 2111 | 2085 | 0.003 | 0.010 | -0.104 | 0.082 | -0.149 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| WNK2 |   | 7 | 256 | 1 | 47 | -0.127 | 0.029 | undef | 0.201 | undef |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ATPIF1 |   | 2 | 743 | 1010 | 1007 | 0.102 | 0.164 | 0.165 | undef | -0.031 |
|---|
| GDPD3 |   | 2 | 743 | 179 | 219 | 0.173 | -0.012 | 0.036 | undef | 0.163 |
|---|
| PSPC1 |   | 6 | 301 | 991 | 964 | 0.203 | -0.289 | 0.298 | 0.148 | -0.107 |
|---|
| PFKM |   | 6 | 301 | 83 | 105 | -0.030 | -0.079 | 0.293 | 0.023 | 0.118 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| BCAR3 |   | 2 | 743 | 2111 | 2085 | 0.271 | 0.123 | 0.259 | -0.027 | 0.066 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| ITPR1 |   | 4 | 440 | 1576 | 1543 | -0.096 | 0.077 | 0.015 | undef | 0.008 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| KIF21A |   | 1 | 1195 | 2111 | 2118 | 0.194 | -0.025 | undef | undef | undef |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| CDH2 |   | 10 | 167 | 727 | 702 | 0.169 | 0.109 | -0.016 | 0.123 | 0.155 |
|---|
| TOB1 |   | 6 | 301 | 525 | 517 | 0.055 | 0.097 | -0.069 | 0.107 | -0.081 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6429 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.342E-09 | 3.279E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.343E-08 | 5.305E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 7.513E-08 | 6.118E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.004E-07 | 6.131E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.15E-07 | 5.618E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.171E-07 | 4.769E-05 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.798E-07 | 6.276E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.798E-07 | 5.491E-05 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 2.665E-07 | 7.234E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.917E-07 | 7.126E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 5.853E-07 | 1.3E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.353E-10 | 2.01E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.93E-08 | 5.93E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.334E-08 | 6.684E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.164E-07 | 6.999E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.329E-07 | 6.396E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.332E-07 | 5.343E-05 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 2.04E-07 | 7.012E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.04E-07 | 6.135E-05 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 3.023E-07 | 8.081E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.235E-07 | 7.783E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.312E-07 | 1.381E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.224E-08 | 1.662E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.019E-07 | 1.172E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.729E-07 | 2.092E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 2.767E-07 | 1.591E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.767E-07 | 1.273E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.325E-07 | 1.658E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 4.325E-07 | 1.421E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.733E-07 | 1.361E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.15E-06 | 2.938E-04 |
|---|
| interphase | GO:0051325 |  | 1.344E-06 | 3.092E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.415E-06 | 2.959E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 1.569E-07 | 2.891E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 3.302E-07 | 3.043E-04 |
|---|
| regulation of cell motion | GO:0051270 |  | 3.302E-07 | 2.028E-04 |
|---|
| thymus development | GO:0048538 |  | 3.881E-06 | 1.788E-03 |
|---|
| negative regulation of BMP signaling pathway | GO:0030514 |  | 3.881E-06 | 1.431E-03 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 3.945E-06 | 1.212E-03 |
|---|
| cytoskeleton-dependent intracellular transport | GO:0030705 |  | 4.91E-06 | 1.293E-03 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 4.91E-06 | 1.131E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.039E-06 | 1.237E-03 |
|---|
| positive regulation of growth | GO:0045927 |  | 8.858E-06 | 1.632E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.191E-06 | 1.54E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.224E-08 | 1.662E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.019E-07 | 1.172E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.729E-07 | 2.092E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 2.767E-07 | 1.591E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.767E-07 | 1.273E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.325E-07 | 1.658E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 4.325E-07 | 1.421E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.733E-07 | 1.361E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.15E-06 | 2.938E-04 |
|---|
| interphase | GO:0051325 |  | 1.344E-06 | 3.092E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.415E-06 | 2.959E-04 |
|---|



