Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6418
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7559 | 3.390e-03 | 5.460e-04 | 7.654e-01 | 1.417e-06 |
|---|
| Loi | 0.2296 | 8.133e-02 | 1.164e-02 | 4.308e-01 | 4.079e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.574e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.501e-03 | 0.000e+00 |
|---|
| Wang | 0.2607 | 2.793e-03 | 4.486e-02 | 4.012e-01 | 5.026e-05 |
|---|
Expression data for subnetwork 6418 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
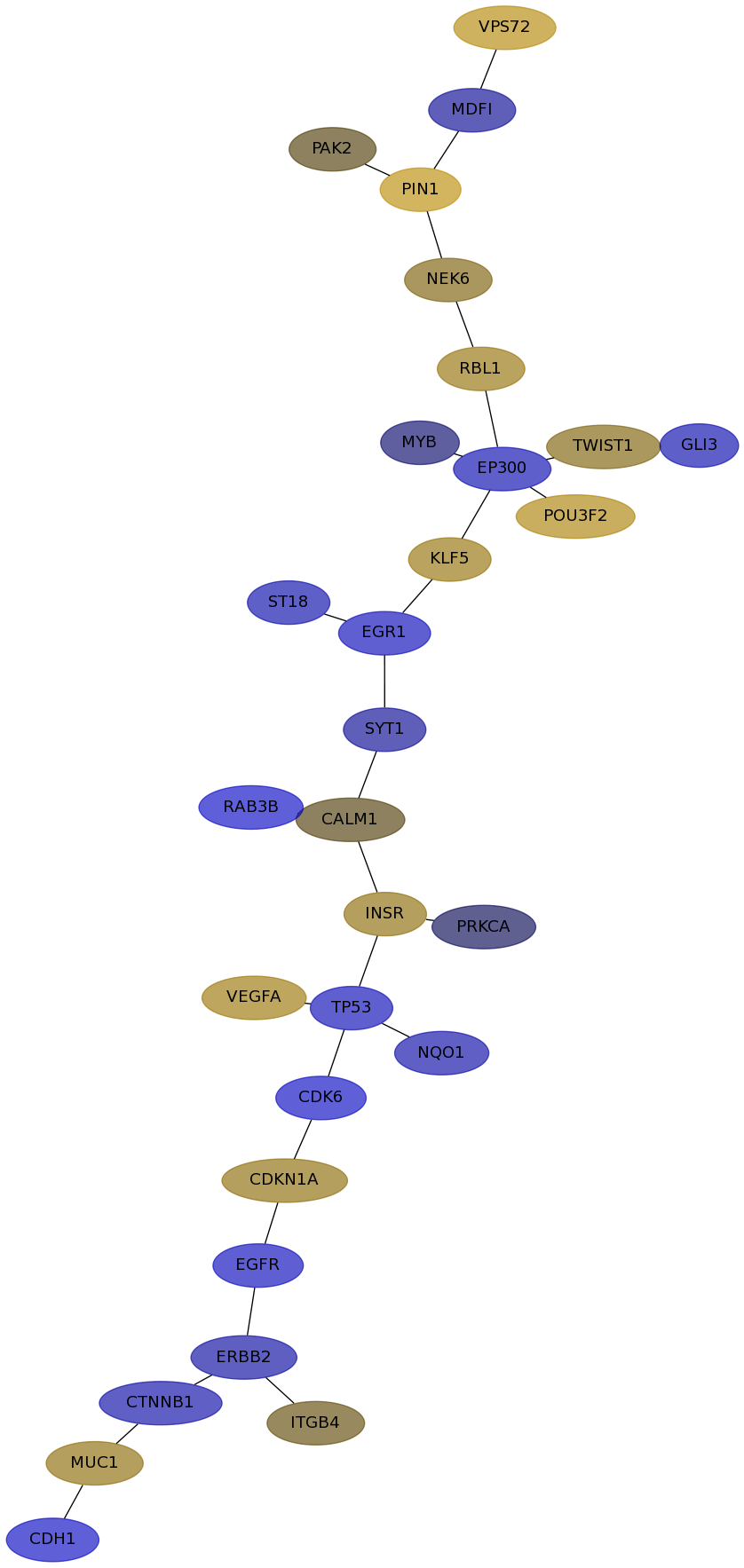
Score for each gene in subnetwork 6418 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| RAB3B |   | 2 | 743 | 1190 | 1189 | -0.249 | -0.034 | -0.132 | 0.125 | 0.116 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| CALM1 |   | 6 | 301 | 614 | 602 | 0.014 | 0.021 | 0.192 | 0.221 | 0.247 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| NEK6 |   | 3 | 557 | 842 | 834 | 0.041 | -0.049 | undef | 0.271 | undef |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| VPS72 |   | 6 | 301 | 318 | 330 | 0.164 | -0.182 | 0.095 | 0.151 | 0.080 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
GO Enrichment output for subnetwork 6418 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 6.773E-09 | 1.655E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.644E-08 | 3.23E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.609E-07 | 1.31E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.825E-07 | 2.336E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 4.621E-07 | 2.258E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.533E-07 | 2.253E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 6.573E-07 | 2.294E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 7.057E-07 | 2.155E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 7.752E-07 | 2.104E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 9.633E-07 | 2.353E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.057E-06 | 2.348E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.093E-09 | 1.947E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.159E-08 | 3.8E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.997E-07 | 1.602E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.744E-07 | 2.252E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 4.566E-07 | 2.197E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 6.603E-07 | 2.648E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.603E-07 | 2.27E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.081E-07 | 2.43E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.629E-07 | 2.307E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.25E-07 | 2.225E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.083E-06 | 2.37E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.931E-09 | 2.284E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.645E-08 | 5.341E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.847E-07 | 2.182E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.214E-07 | 2.998E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 6.658E-07 | 3.063E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 8.203E-07 | 3.144E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.999E-07 | 3.285E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 9.999E-07 | 2.875E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.207E-06 | 3.085E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.472E-06 | 3.385E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.715E-06 | 3.587E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.491E-08 | 2.748E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.777E-07 | 2.559E-04 |
|---|
| regulation of protein import into nucleus | GO:0042306 |  | 3.713E-07 | 2.281E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.786E-07 | 3.588E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.076E-06 | 3.966E-04 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 1.259E-06 | 3.868E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.45E-06 | 3.819E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.914E-06 | 4.409E-04 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 2.035E-06 | 4.167E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.547E-06 | 6.538E-04 |
|---|
| lung development | GO:0030324 |  | 3.97E-06 | 6.651E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.931E-09 | 2.284E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.645E-08 | 5.341E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.847E-07 | 2.182E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.214E-07 | 2.998E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 6.658E-07 | 3.063E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 8.203E-07 | 3.144E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.999E-07 | 3.285E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 9.999E-07 | 2.875E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.207E-06 | 3.085E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.472E-06 | 3.385E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.715E-06 | 3.587E-04 |
|---|



