Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6408
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7561 | 3.375e-03 | 5.430e-04 | 7.648e-01 | 1.402e-06 |
|---|
| Loi | 0.2296 | 8.140e-02 | 1.166e-02 | 4.309e-01 | 4.090e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.583e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.495e-03 | 0.000e+00 |
|---|
| Wang | 0.2606 | 2.819e-03 | 4.509e-02 | 4.022e-01 | 5.113e-05 |
|---|
Expression data for subnetwork 6408 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
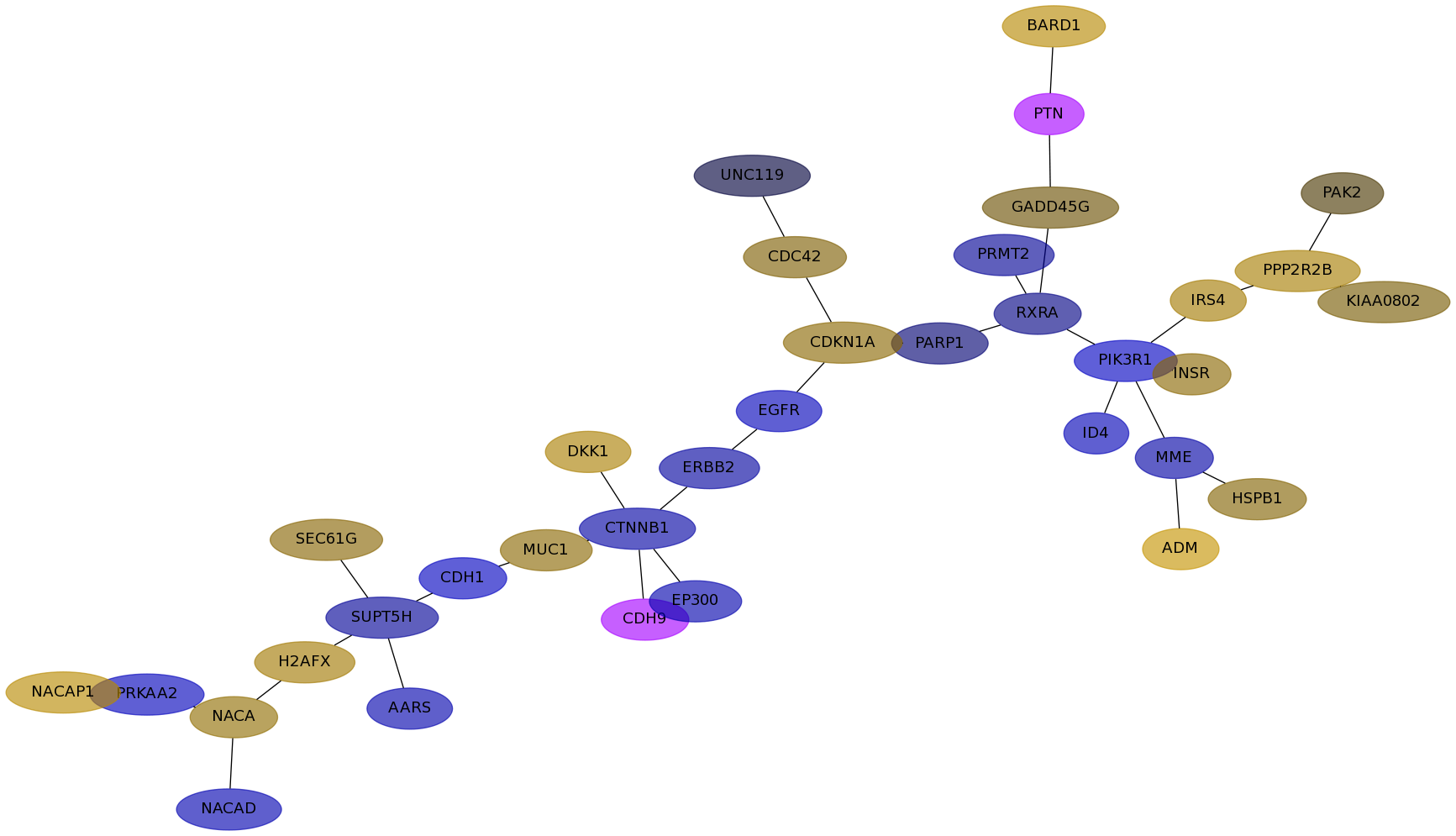
Score for each gene in subnetwork 6408 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PRKAA2 |   | 6 | 301 | 1165 | 1138 | -0.204 | 0.148 | 0.086 | 0.250 | -0.078 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NACAP1 |   | 2 | 743 | 1323 | 1313 | 0.186 | 0.227 | 0.076 | undef | 0.244 |
|---|
| SEC61G |   | 1 | 1195 | 2206 | 2210 | 0.058 | 0.019 | -0.211 | 0.268 | 0.098 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| NACA |   | 5 | 360 | 1165 | 1143 | 0.075 | 0.158 | 0.046 | -0.135 | 0.252 |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| NACAD |   | 3 | 557 | 1323 | 1309 | -0.159 | 0.096 | 0.093 | 0.155 | -0.078 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| AARS |   | 2 | 743 | 1564 | 1549 | -0.148 | 0.030 | -0.164 | 0.177 | 0.035 |
|---|
| CDH9 |   | 2 | 743 | 1396 | 1391 | undef | -0.013 | 0.162 | 0.199 | 0.193 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SUPT5H |   | 6 | 301 | 1323 | 1281 | -0.085 | 0.007 | -0.298 | 0.002 | -0.068 |
|---|
| PRMT2 |   | 5 | 360 | 957 | 938 | -0.081 | 0.046 | 0.089 | 0.235 | -0.128 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| DKK1 |   | 4 | 440 | 236 | 262 | 0.134 | 0.091 | -0.124 | 0.065 | 0.129 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| KIAA0802 |   | 3 | 557 | 2046 | 2009 | 0.041 | -0.123 | 0.123 | 0.059 | 0.037 |
|---|
| H2AFX |   | 7 | 256 | 179 | 201 | 0.111 | -0.072 | 0.121 | 0.165 | -0.010 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| MME |   | 5 | 360 | 1047 | 1021 | -0.121 | 0.020 | 0.088 | -0.002 | -0.130 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
GO Enrichment output for subnetwork 6408 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.443E-07 | 5.969E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.443E-07 | 2.984E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.205E-07 | 3.424E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.818E-07 | 4.164E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.818E-07 | 3.331E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.818E-07 | 2.776E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.021E-06 | 3.563E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.999E-06 | 6.103E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.999E-06 | 5.425E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.66E-06 | 6.499E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.185E-06 | 9.295E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.324E-07 | 7.996E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.324E-07 | 3.998E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.998E-07 | 4.009E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.805E-07 | 3.492E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 5.805E-07 | 2.794E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.271E-07 | 3.718E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 9.271E-07 | 3.187E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.979E-06 | 5.953E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.716E-06 | 7.262E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.615E-06 | 8.698E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.254E-06 | 1.368E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 4.655E-07 | 1.071E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.655E-07 | 5.353E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.12E-07 | 4.692E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.286E-07 | 5.339E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.621E-06 | 7.456E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.621E-06 | 6.213E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.586E-06 | 8.498E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.87E-06 | 1.113E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.514E-06 | 1.409E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 7.561E-06 | 1.739E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.006E-05 | 2.102E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.85E-08 | 7.095E-05 |
|---|
| heart development | GO:0007507 |  | 5.018E-07 | 4.624E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 5.372E-07 | 3.301E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 6.862E-06 | 3.162E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.184E-05 | 4.363E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.185E-05 | 3.639E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.503E-05 | 3.957E-03 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 2.301E-05 | 5.301E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.574E-05 | 5.271E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 8.398E-05 | 0.01547801 |
|---|
| striated muscle tissue development | GO:0014706 |  | 8.91E-05 | 0.01492771 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 4.655E-07 | 1.071E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.655E-07 | 5.353E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.12E-07 | 4.692E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.286E-07 | 5.339E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.621E-06 | 7.456E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.621E-06 | 6.213E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.586E-06 | 8.498E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.87E-06 | 1.113E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.514E-06 | 1.409E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 7.561E-06 | 1.739E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.006E-05 | 2.102E-03 |
|---|



