Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6389
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7567 | 3.343e-03 | 5.360e-04 | 7.633e-01 | 1.368e-06 |
|---|
| Loi | 0.2294 | 8.165e-02 | 1.173e-02 | 4.316e-01 | 4.133e-04 |
|---|
| Schmidt | 0.6724 | 0.000e+00 | 0.000e+00 | 4.602e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.496e-03 | 0.000e+00 |
|---|
| Wang | 0.2604 | 2.836e-03 | 4.524e-02 | 4.029e-01 | 5.169e-05 |
|---|
Expression data for subnetwork 6389 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
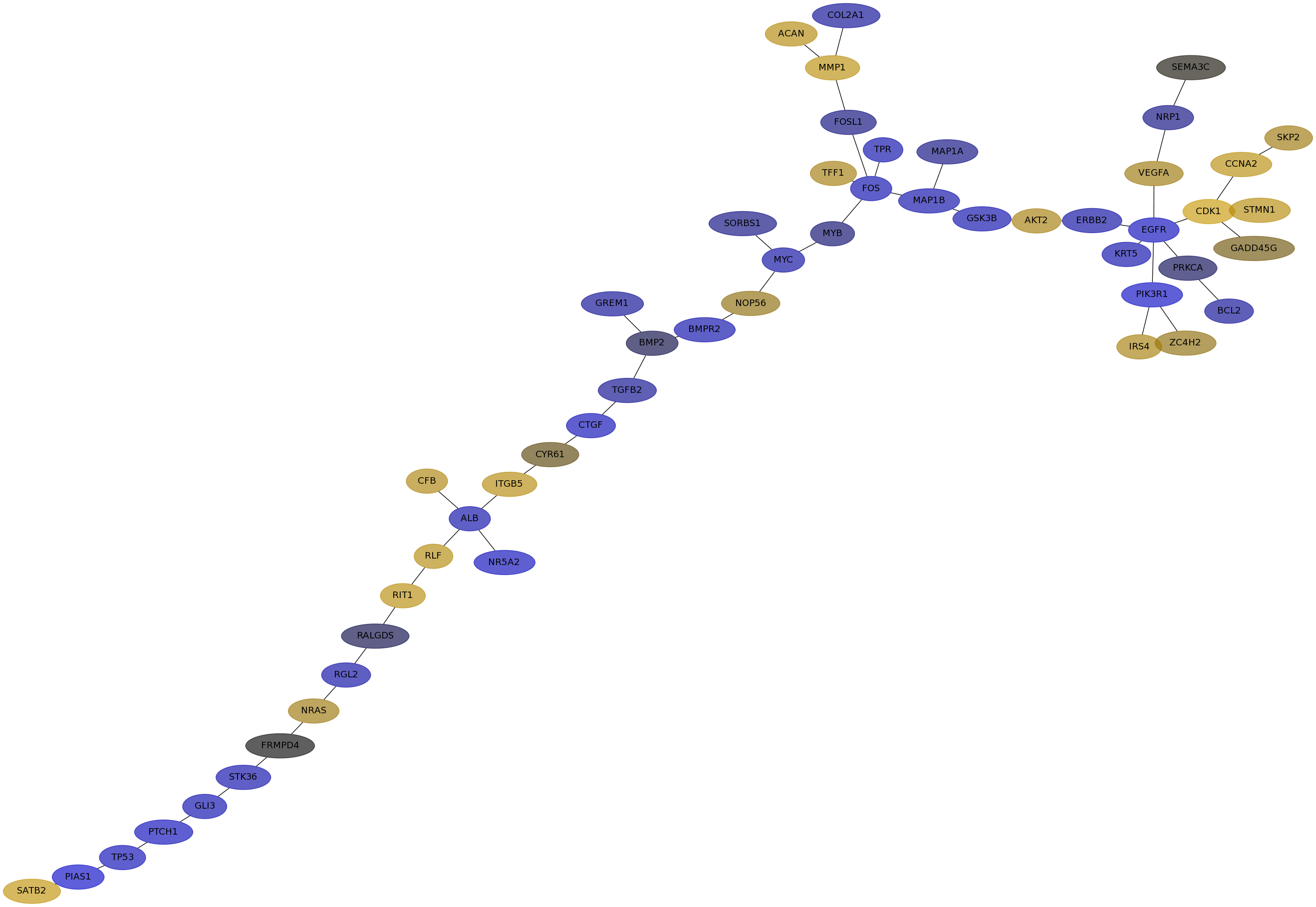
Score for each gene in subnetwork 6389 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| BMPR2 |   | 5 | 360 | 1323 | 1292 | -0.132 | 0.081 | -0.029 | 0.356 | -0.021 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| RALGDS |   | 5 | 360 | 2024 | 1972 | -0.011 | 0.199 | 0.033 | undef | -0.009 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| STK36 |   | 1 | 1195 | 2090 | 2095 | -0.125 | 0.146 | undef | undef | undef |
|---|
| TGFB2 |   | 7 | 256 | 525 | 515 | -0.063 | 0.244 | 0.226 | 0.079 | 0.015 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| ITGB5 |   | 4 | 440 | 412 | 419 | 0.163 | 0.208 | 0.081 | 0.278 | 0.148 |
|---|
| RLF |   | 2 | 743 | 179 | 219 | 0.158 | 0.178 | -0.013 | -0.043 | -0.046 |
|---|
| STMN1 |   | 4 | 440 | 1461 | 1437 | 0.168 | -0.033 | 0.198 | 0.105 | 0.062 |
|---|
| NOP56 |   | 3 | 557 | 1450 | 1430 | 0.062 | 0.036 | 0.030 | undef | 0.011 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| PIAS1 |   | 2 | 743 | 2090 | 2082 | -0.270 | 0.096 | 0.157 | -0.213 | -0.083 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CYR61 |   | 4 | 440 | 991 | 978 | 0.018 | 0.206 | 0.203 | 0.092 | 0.234 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| COL2A1 |   | 2 | 743 | 525 | 547 | -0.077 | -0.000 | 0.174 | -0.137 | 0.173 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| NRAS |   | 2 | 743 | 2090 | 2082 | 0.090 | -0.211 | -0.054 | 0.076 | 0.021 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| MMP1 |   | 11 | 148 | 366 | 360 | 0.192 | 0.014 | 0.036 | 0.044 | 0.260 |
|---|
| PTCH1 |   | 5 | 360 | 1190 | 1157 | -0.184 | 0.047 | 0.121 | -0.055 | 0.004 |
|---|
| SATB2 |   | 1 | 1195 | 2090 | 2095 | 0.222 | 0.233 | 0.166 | 0.025 | 0.050 |
|---|
| RIT1 |   | 1 | 1195 | 2090 | 2095 | 0.179 | 0.027 | 0.048 | 0.186 | 0.009 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NR5A2 |   | 1 | 1195 | 2090 | 2095 | -0.186 | -0.150 | 0.162 | 0.096 | 0.186 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TFF1 |   | 7 | 256 | 957 | 928 | 0.106 | 0.022 | 0.088 | -0.033 | -0.244 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| ALB |   | 3 | 557 | 412 | 430 | -0.122 | 0.218 | 0.080 | 0.018 | 0.038 |
|---|
| FRMPD4 |   | 1 | 1195 | 2090 | 2095 | 0.000 | 0.051 | -0.088 | 0.159 | -0.068 |
|---|
| ACAN |   | 5 | 360 | 412 | 416 | 0.156 | 0.132 | -0.024 | -0.171 | -0.083 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| SEMA3C |   | 2 | 743 | 2090 | 2082 | 0.002 | 0.120 | 0.128 | 0.227 | -0.154 |
|---|
| RGL2 |   | 1 | 1195 | 2090 | 2095 | -0.107 | 0.227 | -0.146 | 0.070 | -0.002 |
|---|
| CFB |   | 1 | 1195 | 2090 | 2095 | 0.138 | 0.046 | 0.109 | -0.061 | -0.149 |
|---|
| NRP1 |   | 8 | 222 | 1222 | 1186 | -0.043 | 0.025 | 0.201 | 0.029 | 0.071 |
|---|
| FOSL1 |   | 11 | 148 | 366 | 360 | -0.040 | 0.164 | 0.010 | 0.128 | 0.005 |
|---|
GO Enrichment output for subnetwork 6389 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 9.875E-10 | 2.412E-06 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.775E-09 | 3.389E-06 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 2.838E-09 | 2.311E-06 |
|---|
| tube morphogenesis | GO:0035239 |  | 5.426E-09 | 3.314E-06 |
|---|
| ureteric bud development | GO:0001657 |  | 3.549E-08 | 1.734E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.198E-08 | 1.709E-05 |
|---|
| cartilage development | GO:0051216 |  | 1.164E-07 | 4.062E-05 |
|---|
| epithelial tube morphogenesis | GO:0060562 |  | 1.305E-07 | 3.984E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.633E-07 | 4.432E-05 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 1.814E-07 | 4.431E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.016E-07 | 4.477E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 1.427E-09 | 3.435E-06 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 3.637E-09 | 4.375E-06 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 4.098E-09 | 3.286E-06 |
|---|
| tube morphogenesis | GO:0035239 |  | 6.22E-09 | 3.741E-06 |
|---|
| ureteric bud development | GO:0001657 |  | 4.645E-08 | 2.235E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 5.223E-08 | 2.095E-05 |
|---|
| epithelial tube morphogenesis | GO:0060562 |  | 1.261E-07 | 4.335E-05 |
|---|
| cartilage development | GO:0051216 |  | 1.42E-07 | 4.272E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.03E-07 | 5.427E-05 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.485E-07 | 5.978E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.761E-07 | 6.039E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 4.887E-09 | 1.124E-05 |
|---|
| tube morphogenesis | GO:0035239 |  | 1.166E-08 | 1.341E-05 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.297E-08 | 9.946E-06 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 1.504E-08 | 8.649E-06 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.123E-08 | 3.736E-05 |
|---|
| ureteric bud development | GO:0001657 |  | 1.643E-07 | 6.299E-05 |
|---|
| epithelial tube morphogenesis | GO:0060562 |  | 1.954E-07 | 6.421E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.777E-07 | 1.086E-04 |
|---|
| cartilage development | GO:0051216 |  | 4.355E-07 | 1.113E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 7.014E-07 | 1.613E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 7.853E-07 | 1.642E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 1.125E-08 | 2.073E-05 |
|---|
| tube development | GO:0035295 |  | 1.472E-08 | 1.357E-05 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 2.119E-08 | 1.302E-05 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.209E-07 | 5.57E-05 |
|---|
| tube morphogenesis | GO:0035239 |  | 2.064E-07 | 7.607E-05 |
|---|
| cell aging | GO:0007569 |  | 6.746E-07 | 2.072E-04 |
|---|
| ureteric bud development | GO:0001657 |  | 6.746E-07 | 1.776E-04 |
|---|
| regulation of neuron apoptosis | GO:0043523 |  | 1.081E-06 | 2.49E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 1.262E-06 | 2.584E-04 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 1.277E-06 | 2.354E-04 |
|---|
| ectoderm development | GO:0007398 |  | 1.304E-06 | 2.185E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 4.887E-09 | 1.124E-05 |
|---|
| tube morphogenesis | GO:0035239 |  | 1.166E-08 | 1.341E-05 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.297E-08 | 9.946E-06 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 1.504E-08 | 8.649E-06 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.123E-08 | 3.736E-05 |
|---|
| ureteric bud development | GO:0001657 |  | 1.643E-07 | 6.299E-05 |
|---|
| epithelial tube morphogenesis | GO:0060562 |  | 1.954E-07 | 6.421E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.777E-07 | 1.086E-04 |
|---|
| cartilage development | GO:0051216 |  | 4.355E-07 | 1.113E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 7.014E-07 | 1.613E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 7.853E-07 | 1.642E-04 |
|---|



