Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6369
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7572 | 3.310e-03 | 5.290e-04 | 7.619e-01 | 1.334e-06 |
|---|
| Loi | 0.2292 | 8.207e-02 | 1.184e-02 | 4.326e-01 | 4.203e-04 |
|---|
| Schmidt | 0.6720 | 0.000e+00 | 0.000e+00 | 4.642e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.491e-03 | 0.000e+00 |
|---|
| Wang | 0.2604 | 2.846e-03 | 4.533e-02 | 4.033e-01 | 5.203e-05 |
|---|
Expression data for subnetwork 6369 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
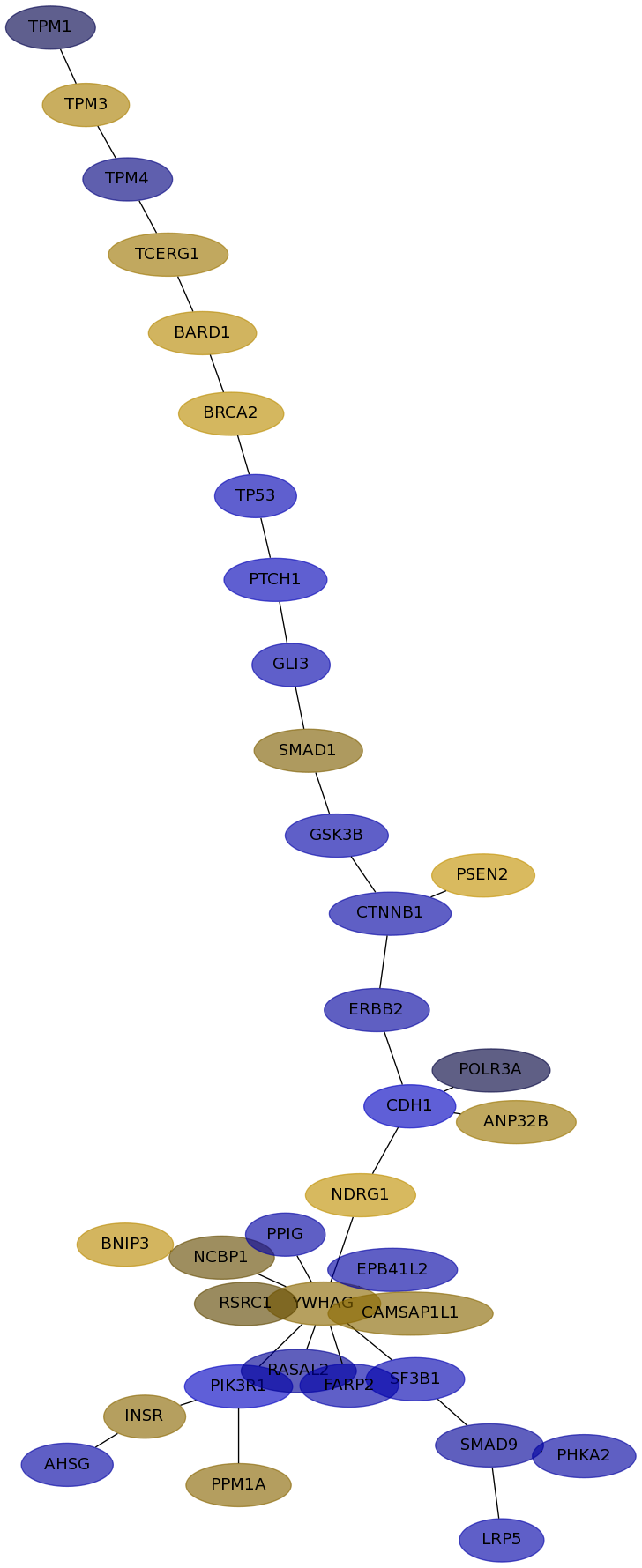
Score for each gene in subnetwork 6369 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| RASAL2 |   | 1 | 1195 | 2239 | 2240 | -0.083 | 0.191 | -0.074 | undef | 0.003 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| NDRG1 |   | 6 | 301 | 658 | 646 | 0.222 | 0.143 | -0.040 | -0.018 | 0.242 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| EPB41L2 |   | 1 | 1195 | 2239 | 2240 | -0.118 | 0.172 | 0.132 | -0.179 | 0.242 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| PPM1A |   | 1 | 1195 | 2239 | 2240 | 0.060 | -0.059 | -0.034 | 0.056 | -0.035 |
|---|
| ANP32B |   | 2 | 743 | 1822 | 1806 | 0.096 | 0.080 | 0.309 | -0.081 | 0.045 |
|---|
| PHKA2 |   | 3 | 557 | 991 | 982 | -0.107 | 0.059 | -0.153 | 0.250 | 0.041 |
|---|
| FARP2 |   | 1 | 1195 | 2239 | 2240 | -0.127 | -0.115 | -0.030 | 0.049 | 0.101 |
|---|
| TPM3 |   | 2 | 743 | 1903 | 1878 | 0.135 | 0.036 | undef | undef | undef |
|---|
| SMAD9 |   | 9 | 196 | 296 | 285 | -0.090 | -0.153 | 0.055 | 0.081 | -0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| TPM4 |   | 1 | 1195 | 2239 | 2240 | -0.048 | 0.161 | 0.034 | 0.005 | -0.078 |
|---|
| PTCH1 |   | 5 | 360 | 1190 | 1157 | -0.184 | 0.047 | 0.121 | -0.055 | 0.004 |
|---|
| CAMSAP1L1 |   | 4 | 440 | 296 | 321 | 0.062 | 0.083 | 0.113 | 0.151 | 0.150 |
|---|
| LRP5 |   | 3 | 557 | 1141 | 1129 | -0.129 | -0.051 | 0.011 | 0.017 | -0.146 |
|---|
| TCERG1 |   | 4 | 440 | 412 | 419 | 0.098 | 0.093 | 0.172 | 0.228 | 0.105 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| AHSG |   | 5 | 360 | 412 | 416 | -0.113 | -0.054 | 0.000 | 0.132 | undef |
|---|
| NCBP1 |   | 3 | 557 | 1294 | 1276 | 0.027 | -0.115 | 0.006 | -0.024 | -0.093 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| SMAD1 |   | 5 | 360 | 682 | 671 | 0.049 | 0.159 | -0.062 | 0.140 | 0.048 |
|---|
| TPM1 |   | 1 | 1195 | 2239 | 2240 | -0.014 | 0.228 | 0.253 | 0.081 | 0.131 |
|---|
GO Enrichment output for subnetwork 6369 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.196E-08 | 2.921E-05 |
|---|
| mammary gland development | GO:0030879 |  | 5.394E-08 | 6.588E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.945E-08 | 5.656E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.11E-07 | 6.782E-05 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.512E-07 | 7.388E-05 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.725E-07 | 7.025E-05 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.595E-07 | 9.057E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.319E-07 | 1.014E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.273E-07 | 1.431E-04 |
|---|
| neuron remodeling | GO:0016322 |  | 5.273E-07 | 1.288E-04 |
|---|
| ER overload response | GO:0006983 |  | 8.421E-07 | 1.87E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.007E-08 | 2.423E-05 |
|---|
| mammary gland development | GO:0030879 |  | 6.653E-08 | 8.004E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.768E-08 | 7.834E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.561E-07 | 9.389E-05 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.867E-07 | 8.984E-05 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.276E-07 | 9.127E-05 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.423E-07 | 1.176E-04 |
|---|
| neuron remodeling | GO:0016322 |  | 3.727E-07 | 1.121E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 4.659E-07 | 1.245E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.509E-07 | 1.566E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.039E-06 | 2.273E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mammary gland development | GO:0030879 |  | 4.085E-08 | 9.395E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.73E-08 | 8.89E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.992E-07 | 1.527E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.024E-07 | 1.164E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.934E-07 | 1.349E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.281E-07 | 1.258E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 4.392E-07 | 1.443E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.854E-07 | 1.683E-04 |
|---|
| ER overload response | GO:0006983 |  | 5.854E-07 | 1.496E-04 |
|---|
| midbrain development | GO:0030901 |  | 5.854E-07 | 1.346E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.632E-06 | 3.412E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.64E-08 | 8.552E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 7.01E-08 | 6.46E-05 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 8.233E-08 | 5.058E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.458E-07 | 6.719E-05 |
|---|
| ER overload response | GO:0006983 |  | 1.336E-06 | 4.923E-04 |
|---|
| regulation of protein transport | GO:0051223 |  | 2.31E-06 | 7.095E-04 |
|---|
| regulation of mRNA processing | GO:0050684 |  | 2.331E-06 | 6.136E-04 |
|---|
| regulation of protein localization | GO:0032880 |  | 3.331E-06 | 7.674E-04 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 5.562E-06 | 1.139E-03 |
|---|
| positive regulation of protein transport | GO:0051222 |  | 5.623E-06 | 1.036E-03 |
|---|
| positive regulation of transport | GO:0051050 |  | 9.27E-06 | 1.553E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mammary gland development | GO:0030879 |  | 4.085E-08 | 9.395E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.73E-08 | 8.89E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.992E-07 | 1.527E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.024E-07 | 1.164E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.934E-07 | 1.349E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.281E-07 | 1.258E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 4.392E-07 | 1.443E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.854E-07 | 1.683E-04 |
|---|
| ER overload response | GO:0006983 |  | 5.854E-07 | 1.496E-04 |
|---|
| midbrain development | GO:0030901 |  | 5.854E-07 | 1.346E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.632E-06 | 3.412E-04 |
|---|



