Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6348
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7574 | 3.300e-03 | 5.270e-04 | 7.614e-01 | 1.324e-06 |
|---|
| Loi | 0.2292 | 8.199e-02 | 1.182e-02 | 4.324e-01 | 4.189e-04 |
|---|
| Schmidt | 0.6719 | 0.000e+00 | 0.000e+00 | 4.661e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.484e-03 | 0.000e+00 |
|---|
| Wang | 0.2603 | 2.860e-03 | 4.545e-02 | 4.039e-01 | 5.251e-05 |
|---|
Expression data for subnetwork 6348 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
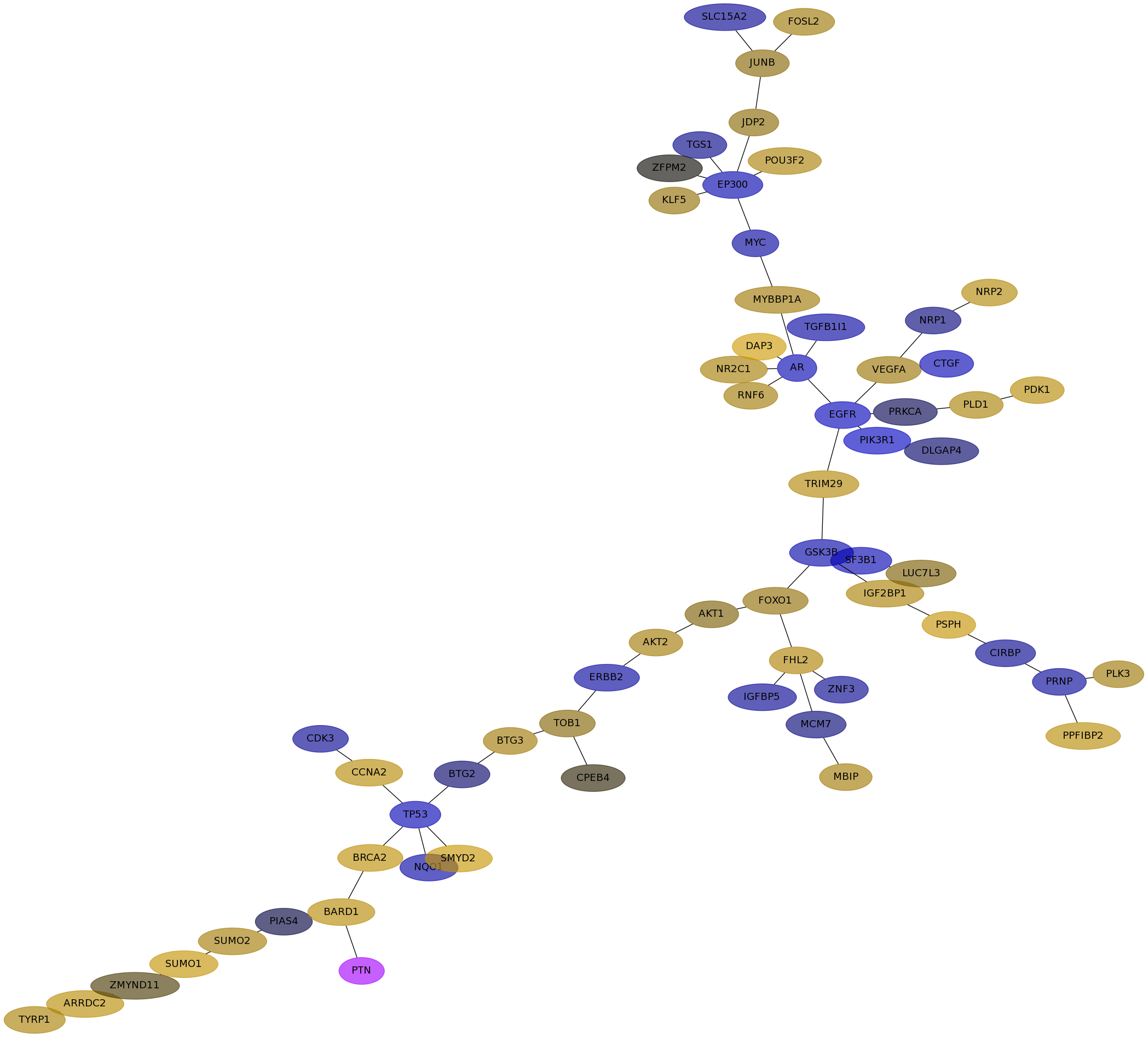
Score for each gene in subnetwork 6348 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| PIAS4 |   | 2 | 743 | 1165 | 1163 | -0.010 | -0.003 | -0.088 | -0.085 | -0.035 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| FOXO1 |   | 3 | 557 | 590 | 598 | 0.072 | 0.094 | -0.021 | -0.147 | 0.221 |
|---|
| CPEB4 |   | 1 | 1195 | 2227 | 2228 | 0.006 | 0.021 | undef | undef | undef |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| JDP2 |   | 4 | 440 | 2053 | 2008 | 0.062 | 0.045 | undef | undef | undef |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| PPFIBP2 |   | 2 | 743 | 2024 | 2002 | 0.188 | 0.320 | -0.112 | 0.053 | 0.013 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| FOSL2 |   | 3 | 557 | 366 | 389 | 0.100 | 0.080 | -0.119 | -0.154 | 0.080 |
|---|
| MCM7 |   | 2 | 743 | 83 | 123 | -0.038 | -0.091 | 0.134 | undef | 0.056 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| NRP2 |   | 1 | 1195 | 2227 | 2228 | 0.160 | 0.033 | -0.004 | 0.026 | 0.002 |
|---|
| CDK3 |   | 7 | 256 | 590 | 570 | -0.076 | 0.189 | -0.041 | -0.089 | 0.082 |
|---|
| SUMO1 |   | 5 | 360 | 1034 | 1004 | 0.245 | -0.007 | 0.144 | 0.083 | 0.118 |
|---|
| PRNP |   | 2 | 743 | 957 | 958 | -0.090 | 0.168 | 0.020 | -0.222 | 0.048 |
|---|
| BTG3 |   | 1 | 1195 | 2227 | 2228 | 0.107 | -0.137 | 0.058 | 0.021 | 0.077 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| ARRDC2 |   | 4 | 440 | 696 | 689 | 0.192 | -0.100 | undef | undef | undef |
|---|
| ZNF3 |   | 1 | 1195 | 2227 | 2228 | -0.067 | 0.147 | 0.056 | undef | -0.027 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| TYRP1 |   | 1 | 1195 | 2227 | 2228 | 0.135 | 0.051 | -0.020 | 0.001 | -0.018 |
|---|
| PLK3 |   | 2 | 743 | 1688 | 1673 | 0.101 | 0.053 | 0.038 | -0.091 | -0.002 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| ZMYND11 |   | 2 | 743 | 696 | 717 | 0.014 | -0.068 | 0.120 | 0.175 | -0.069 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| BTG2 |   | 1 | 1195 | 2227 | 2228 | -0.029 | -0.093 | -0.111 | -0.071 | -0.170 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| CIRBP |   | 5 | 360 | 366 | 379 | -0.069 | 0.208 | 0.110 | 0.049 | -0.045 |
|---|
| ZFPM2 |   | 5 | 360 | 525 | 518 | 0.001 | 0.092 | 0.113 | -0.099 | 0.379 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| SMYD2 |   | 4 | 440 | 1364 | 1330 | 0.260 | 0.068 | 0.179 | 0.014 | 0.191 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| SLC15A2 |   | 4 | 440 | 928 | 912 | -0.076 | 0.109 | -0.068 | 0.050 | -0.123 |
|---|
| MBIP |   | 1 | 1195 | 2227 | 2228 | 0.109 | 0.001 | 0.039 | 0.049 | 0.085 |
|---|
| RNF6 |   | 4 | 440 | 590 | 585 | 0.109 | -0.128 | 0.002 | 0.041 | -0.018 |
|---|
| MYBBP1A |   | 1 | 1195 | 2227 | 2228 | 0.104 | 0.090 | 0.146 | -0.089 | 0.030 |
|---|
| DAP3 |   | 3 | 557 | 1425 | 1409 | 0.306 | -0.088 | -0.047 | 0.166 | 0.081 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| NRP1 |   | 8 | 222 | 1222 | 1186 | -0.043 | 0.025 | 0.201 | 0.029 | 0.071 |
|---|
| TGS1 |   | 5 | 360 | 780 | 767 | -0.060 | -0.044 | 0.232 | undef | 0.269 |
|---|
| TOB1 |   | 6 | 301 | 525 | 517 | 0.055 | 0.097 | -0.069 | 0.107 | -0.081 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
GO Enrichment output for subnetwork 6348 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 9.733E-15 | 2.378E-11 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.214E-08 | 1.003E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.145E-07 | 9.324E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.188E-07 | 1.947E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.924E-06 | 9.401E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 2.611E-06 | 1.063E-03 |
|---|
| negative regulation of axon extension | GO:0030517 |  | 2.611E-06 | 9.114E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.022E-06 | 9.229E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 4.526E-06 | 1.229E-03 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.526E-06 | 1.106E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.553E-06 | 1.011E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 2.073E-14 | 4.987E-11 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.266E-07 | 1.523E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.153E-07 | 1.727E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.907E-07 | 2.951E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 1.815E-06 | 8.735E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 2.307E-06 | 9.251E-04 |
|---|
| negative regulation of axon extension | GO:0030517 |  | 3.615E-06 | 1.243E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.634E-06 | 1.394E-03 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 5.699E-06 | 1.523E-03 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 5.699E-06 | 1.371E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.3E-06 | 1.378E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 7.531E-14 | 1.732E-10 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 1.191E-08 | 1.369E-05 |
|---|
| protein modification by small protein conjugation or removal | GO:0070647 |  | 2.111E-08 | 1.619E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.496E-07 | 8.602E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.073E-07 | 1.414E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.94E-07 | 2.66E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.488E-06 | 4.888E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 4.847E-06 | 1.394E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.199E-06 | 1.584E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.371E-06 | 1.465E-03 |
|---|
| ER overload response | GO:0006983 |  | 6.371E-06 | 1.332E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 2.163E-08 | 3.986E-05 |
|---|
| negative regulation of cell development | GO:0010721 |  | 3.26E-06 | 3.004E-03 |
|---|
| ER overload response | GO:0006983 |  | 4.817E-06 | 2.959E-03 |
|---|
| negative regulation of axonogenesis | GO:0050771 |  | 4.817E-06 | 2.219E-03 |
|---|
| negative regulation of cell projection organization | GO:0031345 |  | 8.392E-06 | 3.093E-03 |
|---|
| regulation of axon extension | GO:0030516 |  | 8.392E-06 | 2.578E-03 |
|---|
| blood vessel morphogenesis | GO:0048514 |  | 1.257E-05 | 3.31E-03 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 1.599E-05 | 3.685E-03 |
|---|
| protein modification by small protein conjugation or removal | GO:0070647 |  | 2.324E-05 | 4.759E-03 |
|---|
| blood vessel development | GO:0001568 |  | 2.573E-05 | 4.743E-03 |
|---|
| vasculature development | GO:0001944 |  | 2.902E-05 | 4.863E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 7.531E-14 | 1.732E-10 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 1.191E-08 | 1.369E-05 |
|---|
| protein modification by small protein conjugation or removal | GO:0070647 |  | 2.111E-08 | 1.619E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.496E-07 | 8.602E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.073E-07 | 1.414E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.94E-07 | 2.66E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.488E-06 | 4.888E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 4.847E-06 | 1.394E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.199E-06 | 1.584E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.371E-06 | 1.465E-03 |
|---|
| ER overload response | GO:0006983 |  | 6.371E-06 | 1.332E-03 |
|---|



