Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6312
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7578 | 3.274e-03 | 5.220e-04 | 7.602e-01 | 1.299e-06 |
|---|
| Loi | 0.2292 | 8.200e-02 | 1.182e-02 | 4.324e-01 | 4.191e-04 |
|---|
| Schmidt | 0.6716 | 0.000e+00 | 0.000e+00 | 4.689e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.482e-03 | 0.000e+00 |
|---|
| Wang | 0.2601 | 2.890e-03 | 4.572e-02 | 4.051e-01 | 5.352e-05 |
|---|
Expression data for subnetwork 6312 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
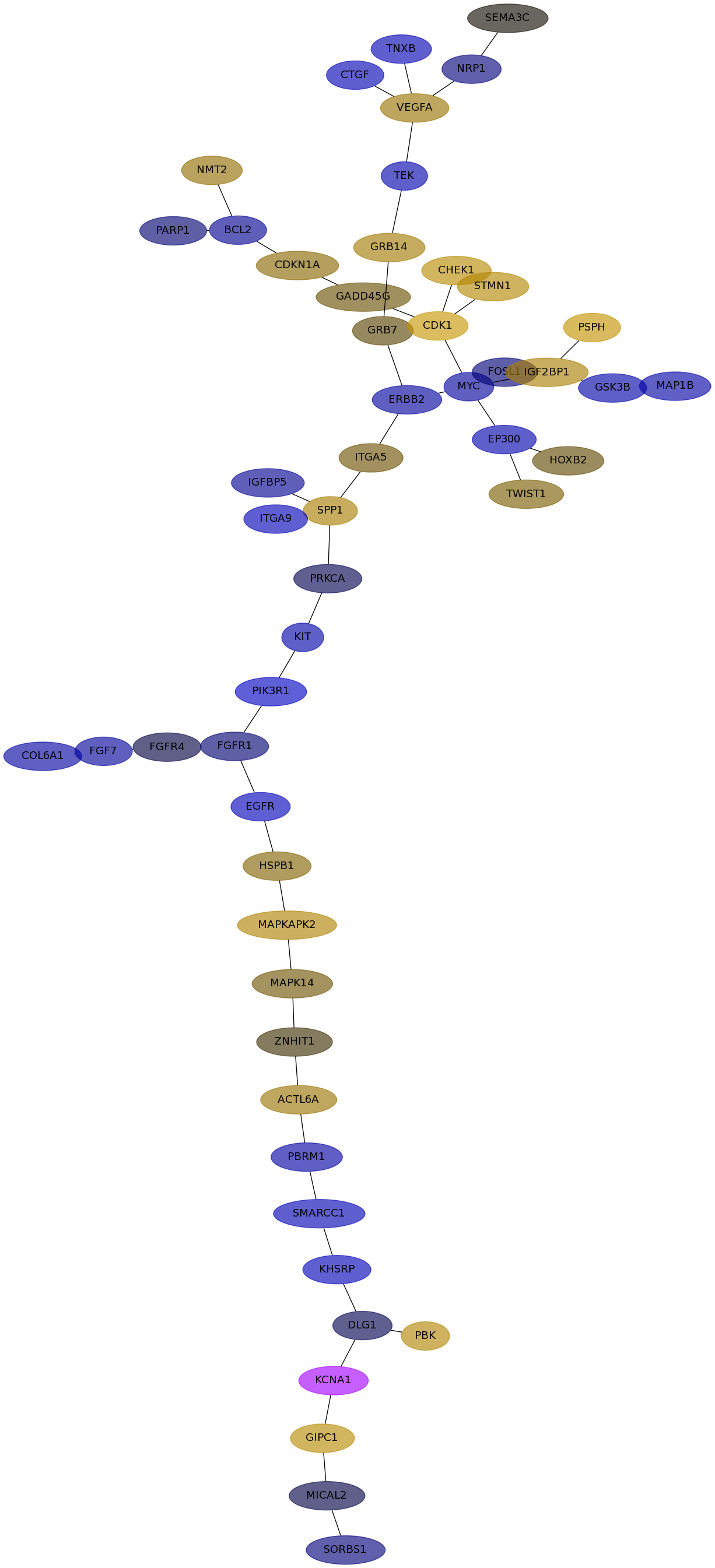
Score for each gene in subnetwork 6312 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| ZNHIT1 |   | 2 | 743 | 318 | 346 | 0.010 | 0.095 | -0.047 | 0.030 | 0.072 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TNXB |   | 4 | 440 | 727 | 723 | -0.169 | -0.076 | 0.160 | 0.182 | 0.173 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| GIPC1 |   | 4 | 440 | 1396 | 1379 | 0.185 | 0.124 | -0.045 | -0.095 | 0.006 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| STMN1 |   | 4 | 440 | 1461 | 1437 | 0.168 | -0.033 | 0.198 | 0.105 | 0.062 |
|---|
| MAPK14 |   | 4 | 440 | 1 | 59 | 0.033 | -0.065 | 0.299 | 0.015 | 0.119 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| COL6A1 |   | 5 | 360 | 780 | 767 | -0.112 | 0.193 | -0.013 | undef | 0.039 |
|---|
| DLG1 |   | 6 | 301 | 457 | 452 | -0.016 | 0.173 | 0.244 | 0.017 | 0.123 |
|---|
| KCNA1 |   | 2 | 743 | 1659 | 1643 | undef | 0.024 | 0.042 | -0.134 | undef |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| ITGA9 |   | 1 | 1195 | 2252 | 2254 | -0.180 | 0.089 | 0.145 | undef | 0.036 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| MICAL2 |   | 4 | 440 | 1659 | 1629 | -0.012 | 0.143 | -0.042 | -0.145 | 0.161 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PBK |   | 2 | 743 | 2207 | 2182 | 0.163 | -0.164 | 0.107 | -0.067 | -0.095 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| PBRM1 |   | 1 | 1195 | 2252 | 2254 | -0.113 | 0.068 | 0.033 | 0.097 | -0.059 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NMT2 |   | 7 | 256 | 906 | 876 | 0.077 | -0.296 | 0.134 | -0.124 | 0.217 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| MAPKAPK2 |   | 3 | 557 | 1681 | 1657 | 0.149 | 0.107 | 0.141 | 0.096 | 0.009 |
|---|
| SMARCC1 |   | 1 | 1195 | 2252 | 2254 | -0.174 | 0.074 | 0.109 | -0.018 | 0.186 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| KHSRP |   | 2 | 743 | 1709 | 1695 | -0.192 | 0.131 | -0.114 | -0.045 | 0.144 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| SEMA3C |   | 2 | 743 | 2090 | 2082 | 0.002 | 0.120 | 0.128 | 0.227 | -0.154 |
|---|
| NRP1 |   | 8 | 222 | 1222 | 1186 | -0.043 | 0.025 | 0.201 | 0.029 | 0.071 |
|---|
| FOSL1 |   | 11 | 148 | 366 | 360 | -0.040 | 0.164 | 0.010 | 0.128 | 0.005 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| ACTL6A |   | 2 | 743 | 1247 | 1238 | 0.095 | -0.069 | 0.193 | undef | 0.040 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
GO Enrichment output for subnetwork 6312 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 5.128E-18 | 1.253E-14 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.105E-12 | 1.349E-09 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 2.445E-12 | 1.991E-09 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 3.981E-12 | 2.431E-09 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 9.707E-12 | 4.743E-09 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 3.942E-11 | 1.605E-08 |
|---|
| extracellular matrix organization | GO:0030198 |  | 6.795E-10 | 2.371E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.835E-09 | 8.658E-07 |
|---|
| aging | GO:0007568 |  | 6.827E-09 | 1.853E-06 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.099E-08 | 2.684E-06 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.422E-08 | 9.821E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.44E-15 | 3.464E-12 |
|---|
| collagen metabolic process | GO:0032963 |  | 9.658E-11 | 1.162E-07 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 1.953E-10 | 1.566E-07 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 6.613E-10 | 3.978E-07 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 1.316E-08 | 6.332E-06 |
|---|
| extracellular matrix organization | GO:0030198 |  | 2.498E-08 | 1.002E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 5.535E-08 | 1.902E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 9.959E-08 | 2.995E-05 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.502E-07 | 4.015E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.151E-07 | 5.175E-05 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 9.765E-07 | 2.136E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 9.397E-15 | 2.161E-11 |
|---|
| collagen metabolic process | GO:0032963 |  | 4.783E-10 | 5.5E-07 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 9.886E-10 | 7.579E-07 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 3.454E-09 | 1.986E-06 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 5.34E-08 | 2.456E-05 |
|---|
| extracellular matrix organization | GO:0030198 |  | 8.774E-08 | 3.363E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.111E-08 | 2.994E-05 |
|---|
| extracellular structure organization | GO:0043062 |  | 1.632E-07 | 4.692E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.029E-07 | 7.74E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.235E-07 | 9.74E-05 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 4.642E-07 | 9.706E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 2.006E-07 | 3.698E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.69E-07 | 4.322E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 9.392E-07 | 5.77E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.208E-06 | 1.017E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 2.208E-06 | 8.139E-04 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 2.238E-06 | 6.874E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 2.837E-06 | 7.469E-04 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 3.598E-06 | 8.29E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 3.598E-06 | 7.368E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 3.598E-06 | 6.632E-04 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 3.729E-06 | 6.247E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 9.397E-15 | 2.161E-11 |
|---|
| collagen metabolic process | GO:0032963 |  | 4.783E-10 | 5.5E-07 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 9.886E-10 | 7.579E-07 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 3.454E-09 | 1.986E-06 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 5.34E-08 | 2.456E-05 |
|---|
| extracellular matrix organization | GO:0030198 |  | 8.774E-08 | 3.363E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.111E-08 | 2.994E-05 |
|---|
| extracellular structure organization | GO:0043062 |  | 1.632E-07 | 4.692E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.029E-07 | 7.74E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.235E-07 | 9.74E-05 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 4.642E-07 | 9.706E-05 |
|---|



