Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6291
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7583 | 3.246e-03 | 5.160e-04 | 7.589e-01 | 1.271e-06 |
|---|
| Loi | 0.2292 | 8.204e-02 | 1.183e-02 | 4.325e-01 | 4.198e-04 |
|---|
| Schmidt | 0.6714 | 0.000e+00 | 0.000e+00 | 4.712e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.487e-03 | 0.000e+00 |
|---|
| Wang | 0.2598 | 2.923e-03 | 4.601e-02 | 4.064e-01 | 5.465e-05 |
|---|
Expression data for subnetwork 6291 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
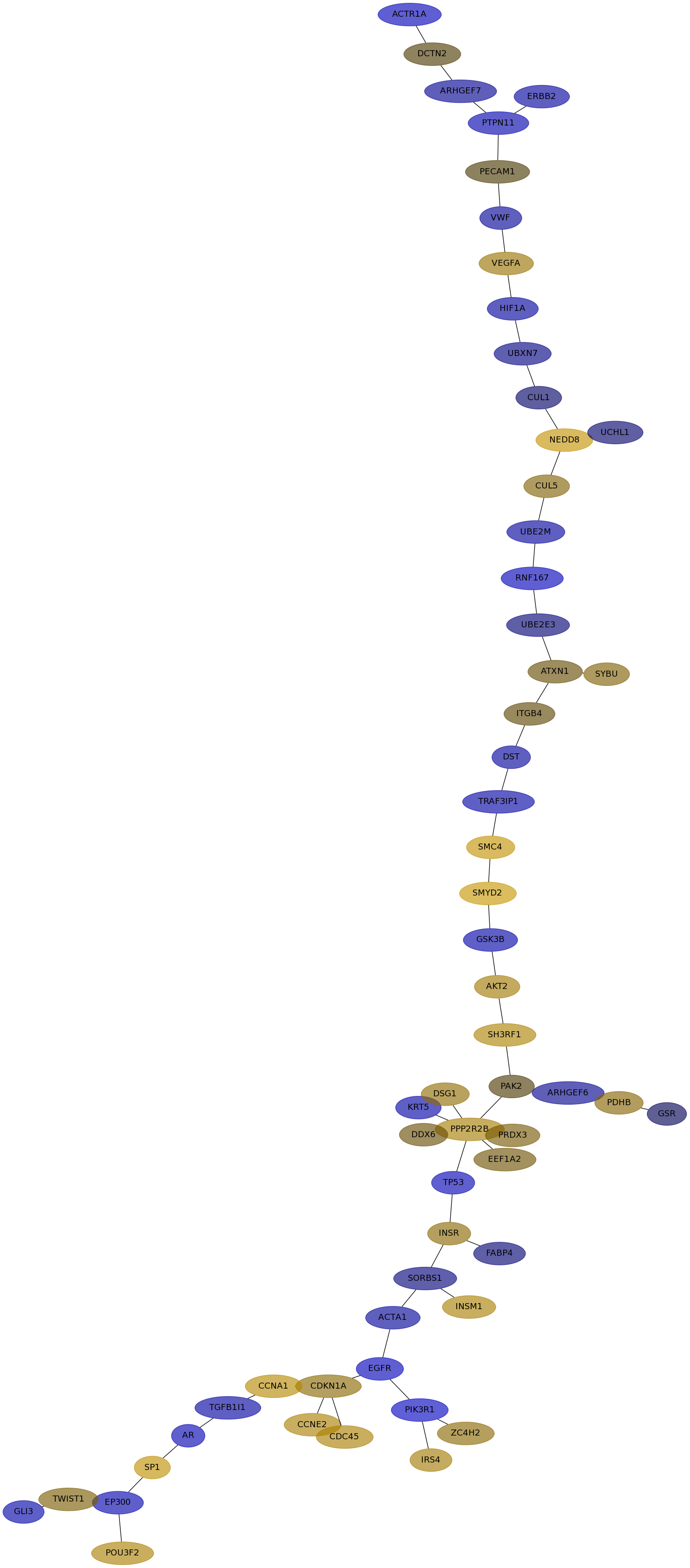
Score for each gene in subnetwork 6291 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CUL1 |   | 1 | 1195 | 2266 | 2267 | -0.028 | -0.216 | -0.072 | 0.027 | -0.041 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PECAM1 |   | 4 | 440 | 83 | 112 | 0.014 | -0.161 | 0.040 | -0.105 | -0.106 |
|---|
| ACTR1A |   | 2 | 743 | 2255 | 2227 | -0.184 | 0.178 | 0.121 | 0.197 | 0.100 |
|---|
| UBXN7 |   | 3 | 557 | 727 | 728 | -0.055 | 0.153 | 0.224 | 0.167 | 0.175 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| CUL5 |   | 3 | 557 | 1735 | 1718 | 0.051 | 0.187 | 0.221 | 0.017 | 0.216 |
|---|
| GSR |   | 3 | 557 | 1222 | 1210 | -0.018 | -0.011 | -0.194 | 0.202 | -0.126 |
|---|
| DDX6 |   | 4 | 440 | 1688 | 1656 | 0.028 | 0.162 | 0.157 | -0.035 | 0.084 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| RNF167 |   | 1 | 1195 | 2266 | 2267 | -0.197 | -0.063 | 0.172 | undef | 0.058 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| UBE2M |   | 1 | 1195 | 2266 | 2267 | -0.099 | -0.123 | -0.231 | 0.047 | -0.152 |
|---|
| UCHL1 |   | 2 | 743 | 1650 | 1635 | -0.031 | -0.164 | 0.135 | -0.011 | 0.024 |
|---|
| SMC4 |   | 3 | 557 | 83 | 117 | 0.247 | -0.111 | 0.214 | 0.072 | 0.094 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| SYBU |   | 3 | 557 | 1055 | 1043 | 0.049 | 0.010 | -0.088 | 0.166 | 0.008 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| SMYD2 |   | 4 | 440 | 1364 | 1330 | 0.260 | 0.068 | 0.179 | 0.014 | 0.191 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDC45 |   | 5 | 360 | 318 | 334 | 0.134 | -0.171 | 0.185 | -0.103 | -0.085 |
|---|
| NEDD8 |   | 4 | 440 | 1650 | 1612 | 0.250 | -0.085 | 0.251 | 0.211 | -0.073 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PRDX3 |   | 1 | 1195 | 2266 | 2267 | 0.038 | -0.059 | 0.173 | 0.055 | -0.143 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| SH3RF1 |   | 6 | 301 | 1222 | 1194 | 0.144 | 0.238 | undef | undef | undef |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| ACTA1 |   | 4 | 440 | 83 | 112 | -0.090 | -0.041 | 0.145 | 0.248 | 0.132 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| VWF |   | 5 | 360 | 525 | 518 | -0.091 | 0.072 | 0.152 | 0.127 | 0.100 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| UBE2E3 |   | 2 | 743 | 2188 | 2166 | -0.040 | 0.003 | 0.040 | 0.149 | -0.137 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| FABP4 |   | 6 | 301 | 236 | 239 | -0.036 | -0.028 | 0.227 | 0.072 | 0.107 |
|---|
GO Enrichment output for subnetwork 6291 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.288E-11 | 8.033E-08 |
|---|
| organ growth | GO:0035265 |  | 1.269E-08 | 1.55E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.269E-08 | 1.033E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.657E-08 | 1.012E-05 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.951E-08 | 9.534E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.529E-08 | 1.03E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.009E-08 | 1.05E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.103E-08 | 9.477E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 4.537E-08 | 1.232E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 5.461E-08 | 1.334E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.06E-07 | 2.355E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.915E-11 | 9.418E-08 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.443E-08 | 1.736E-05 |
|---|
| organ growth | GO:0035265 |  | 1.461E-08 | 1.172E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.461E-08 | 8.79E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 2.03E-08 | 9.769E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.912E-08 | 1.168E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.097E-08 | 1.065E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.8E-08 | 1.143E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.223E-08 | 1.396E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 6.684E-08 | 1.608E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.039E-07 | 2.272E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.479E-11 | 8.001E-08 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 5.224E-09 | 6.008E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 3.124E-08 | 2.395E-05 |
|---|
| organ growth | GO:0035265 |  | 3.624E-08 | 2.084E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 3.624E-08 | 1.667E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 3.776E-08 | 1.447E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.536E-08 | 1.49E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 7.217E-08 | 2.075E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 8.258E-08 | 2.11E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.419E-07 | 3.263E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.282E-07 | 4.772E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.432E-06 | 2.638E-03 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.576E-06 | 3.295E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.256E-06 | 3.229E-03 |
|---|
| ER overload response | GO:0006983 |  | 7.115E-06 | 3.278E-03 |
|---|
| regulation of cellular carbohydrate catabolic process | GO:0043471 |  | 7.115E-06 | 2.623E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 7.897E-06 | 2.426E-03 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.239E-05 | 3.261E-03 |
|---|
| positive regulation of carbohydrate metabolic process | GO:0045913 |  | 1.239E-05 | 2.854E-03 |
|---|
| axon cargo transport | GO:0008088 |  | 1.239E-05 | 2.537E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.349E-05 | 2.486E-03 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.349E-05 | 2.26E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.479E-11 | 8.001E-08 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 5.224E-09 | 6.008E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 3.124E-08 | 2.395E-05 |
|---|
| organ growth | GO:0035265 |  | 3.624E-08 | 2.084E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 3.624E-08 | 1.667E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 3.776E-08 | 1.447E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.536E-08 | 1.49E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 7.217E-08 | 2.075E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 8.258E-08 | 2.11E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.419E-07 | 3.263E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.282E-07 | 4.772E-05 |
|---|



