Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6289
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7585 | 3.232e-03 | 5.130e-04 | 7.582e-01 | 1.257e-06 |
|---|
| Loi | 0.2291 | 8.218e-02 | 1.187e-02 | 4.328e-01 | 4.223e-04 |
|---|
| Schmidt | 0.6714 | 0.000e+00 | 0.000e+00 | 4.714e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.487e-03 | 0.000e+00 |
|---|
| Wang | 0.2598 | 2.922e-03 | 4.600e-02 | 4.064e-01 | 5.462e-05 |
|---|
Expression data for subnetwork 6289 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
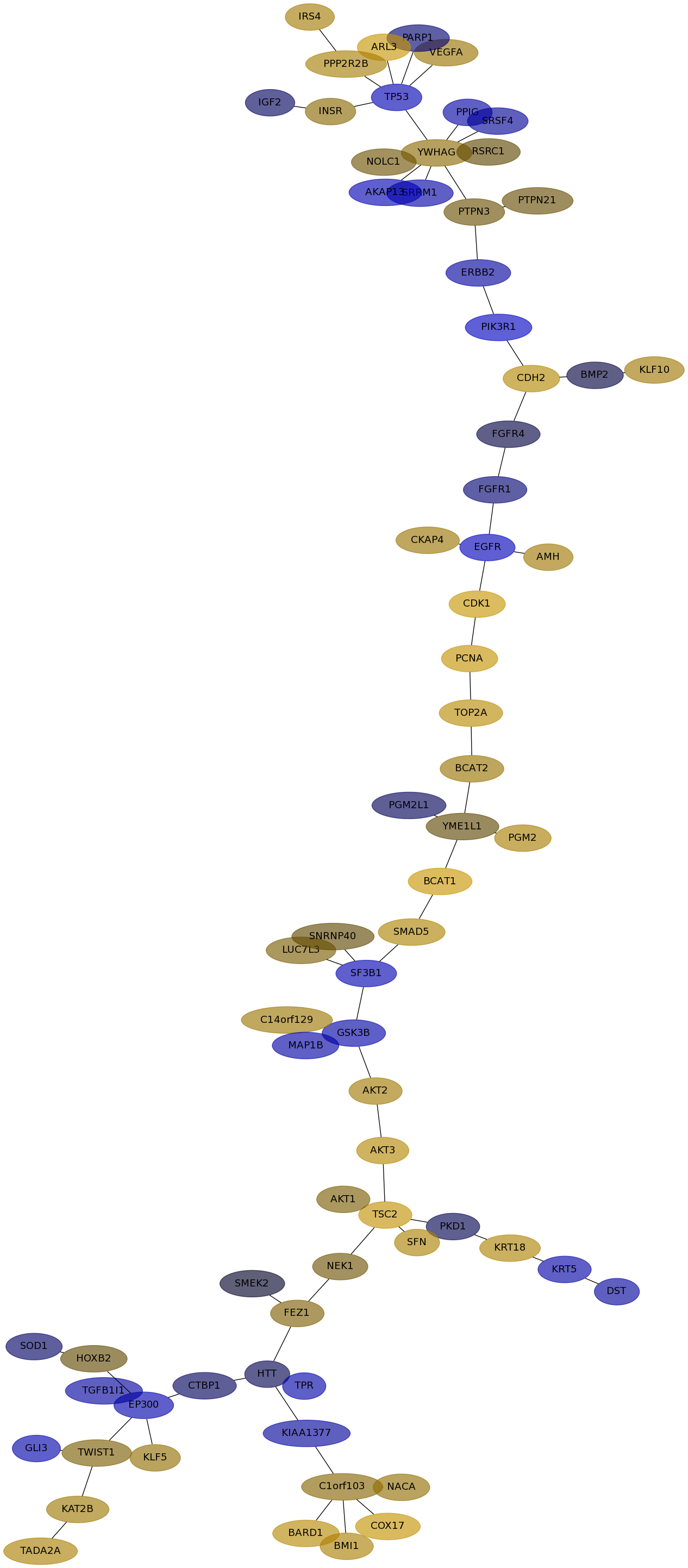
Score for each gene in subnetwork 6289 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SNRNP40 |   | 1 | 1195 | 2129 | 2135 | 0.023 | -0.128 | -0.043 | 0.070 | 0.006 |
|---|
| TSC2 |   | 2 | 743 | 2129 | 2103 | 0.236 | 0.044 | 0.114 | 0.001 | -0.102 |
|---|
| YME1L1 |   | 1 | 1195 | 2129 | 2135 | 0.022 | -0.145 | -0.113 | undef | -0.061 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| PTPN21 |   | 5 | 360 | 1717 | 1677 | 0.027 | 0.177 | 0.214 | 0.205 | 0.119 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| BCAT1 |   | 4 | 440 | 1735 | 1699 | 0.273 | -0.134 | 0.161 | -0.078 | 0.122 |
|---|
| SMAD5 |   | 2 | 743 | 1871 | 1852 | 0.144 | 0.008 | 0.039 | -0.012 | 0.039 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| BCAT2 |   | 3 | 557 | 1510 | 1486 | 0.091 | -0.103 | -0.096 | -0.128 | -0.030 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| SRSF4 |   | 1 | 1195 | 2129 | 2135 | -0.089 | 0.098 | 0.173 | 0.017 | 0.119 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| TADA2A |   | 4 | 440 | 478 | 479 | 0.135 | -0.054 | 0.204 | -0.115 | -0.127 |
|---|
| SOD1 |   | 2 | 743 | 1717 | 1701 | -0.026 | -0.147 | 0.026 | 0.135 | -0.113 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| KLF10 |   | 1 | 1195 | 2129 | 2135 | 0.104 | 0.135 | 0.120 | 0.039 | 0.095 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| AKAP13 |   | 1 | 1195 | 2129 | 2135 | -0.175 | 0.233 | -0.136 | 0.066 | -0.132 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| NOLC1 |   | 4 | 440 | 1047 | 1032 | 0.033 | -0.055 | 0.172 | 0.012 | -0.064 |
|---|
| BMI1 |   | 1 | 1195 | 2129 | 2135 | 0.136 | -0.045 | 0.110 | 0.008 | 0.011 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| PGM2 |   | 1 | 1195 | 2129 | 2135 | 0.127 | -0.013 | undef | -0.226 | undef |
|---|
| NACA |   | 5 | 360 | 1165 | 1143 | 0.075 | 0.158 | 0.046 | -0.135 | 0.252 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| ARL3 |   | 6 | 301 | 83 | 105 | 0.266 | -0.002 | 0.203 | 0.206 | -0.029 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| AMH |   | 10 | 167 | 366 | 362 | 0.103 | 0.056 | 0.113 | -0.187 | 0.301 |
|---|
| SMEK2 |   | 1 | 1195 | 2129 | 2135 | -0.005 | -0.224 | -0.000 | 0.137 | -0.115 |
|---|
| C1orf103 |   | 4 | 440 | 412 | 419 | 0.057 | 0.029 | -0.173 | 0.127 | -0.159 |
|---|
| CKAP4 |   | 6 | 301 | 780 | 764 | 0.093 | 0.152 | -0.167 | 0.315 | -0.017 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| COX17 |   | 2 | 743 | 2129 | 2103 | 0.249 | -0.006 | -0.094 | 0.024 | 0.125 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| AKT3 |   | 5 | 360 | 1083 | 1066 | 0.172 | -0.058 | 0.087 | 0.121 | 0.080 |
|---|
| PGM2L1 |   | 3 | 557 | 412 | 430 | -0.019 | 0.220 | undef | undef | undef |
|---|
| C14orf129 |   | 1 | 1195 | 2129 | 2135 | 0.095 | -0.099 | undef | -0.079 | undef |
|---|
| CDH2 |   | 10 | 167 | 727 | 702 | 0.169 | 0.109 | -0.016 | 0.123 | 0.155 |
|---|
| KRT18 |   | 8 | 222 | 590 | 569 | 0.148 | 0.100 | -0.083 | 0.024 | -0.053 |
|---|
| CTBP1 |   | 3 | 557 | 991 | 982 | -0.022 | -0.021 | -0.298 | 0.196 | -0.143 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
| PKD1 |   | 1 | 1195 | 2129 | 2135 | -0.016 | 0.149 | -0.024 | -0.181 | -0.012 |
|---|
GO Enrichment output for subnetwork 6289 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.298E-07 | 3.171E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.319E-07 | 5.276E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.551E-06 | 2.077E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.027E-06 | 1.849E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.75E-06 | 2.321E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.841E-06 | 2.378E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.411E-06 | 2.237E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 6.411E-06 | 1.958E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 6.411E-06 | 1.74E-03 |
|---|
| regulation of gene-specific transcription | GO:0032583 |  | 6.712E-06 | 1.64E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 7.476E-06 | 1.66E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.659E-07 | 3.991E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.778E-07 | 5.748E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.998E-06 | 2.405E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.016E-06 | 1.814E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.052E-06 | 2.912E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.052E-06 | 2.427E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.712E-06 | 2.651E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 7.712E-06 | 2.319E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 7.712E-06 | 2.062E-03 |
|---|
| regulation of gene-specific transcription | GO:0032583 |  | 9.018E-06 | 2.17E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 9.018E-06 | 1.973E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.539E-07 | 5.84E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.074E-06 | 1.235E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 4.758E-06 | 3.648E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 6.597E-06 | 3.793E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.169E-06 | 3.758E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 9.46E-06 | 3.626E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 9.46E-06 | 3.108E-03 |
|---|
| ER overload response | GO:0006983 |  | 9.46E-06 | 2.72E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.044E-05 | 2.668E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.633E-05 | 3.755E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.646E-05 | 3.442E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ER overload response | GO:0006983 |  | 1.004E-05 | 0.01850859 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.004E-05 | 9.254E-03 |
|---|
| lung development | GO:0030324 |  | 1.065E-05 | 6.54E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 1.389E-05 | 6.399E-03 |
|---|
| respiratory system development | GO:0060541 |  | 1.389E-05 | 5.12E-03 |
|---|
| cranial nerve development | GO:0021545 |  | 1.747E-05 | 5.367E-03 |
|---|
| regionalization | GO:0003002 |  | 1.786E-05 | 4.702E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 2.54E-05 | 5.851E-03 |
|---|
| sex determination | GO:0007530 |  | 2.78E-05 | 5.692E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.78E-05 | 5.123E-03 |
|---|
| nuclear import | GO:0051170 |  | 2.84E-05 | 4.758E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.539E-07 | 5.84E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.074E-06 | 1.235E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 4.758E-06 | 3.648E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 6.597E-06 | 3.793E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.169E-06 | 3.758E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 9.46E-06 | 3.626E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 9.46E-06 | 3.108E-03 |
|---|
| ER overload response | GO:0006983 |  | 9.46E-06 | 2.72E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.044E-05 | 2.668E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.633E-05 | 3.755E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.646E-05 | 3.442E-03 |
|---|



