Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6262
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7593 | 3.189e-03 | 5.050e-04 | 7.561e-01 | 1.218e-06 |
|---|
| Loi | 0.2289 | 8.240e-02 | 1.193e-02 | 4.334e-01 | 4.259e-04 |
|---|
| Schmidt | 0.6711 | 0.000e+00 | 0.000e+00 | 4.743e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.476e-03 | 0.000e+00 |
|---|
| Wang | 0.2596 | 2.959e-03 | 4.632e-02 | 4.078e-01 | 5.590e-05 |
|---|
Expression data for subnetwork 6262 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
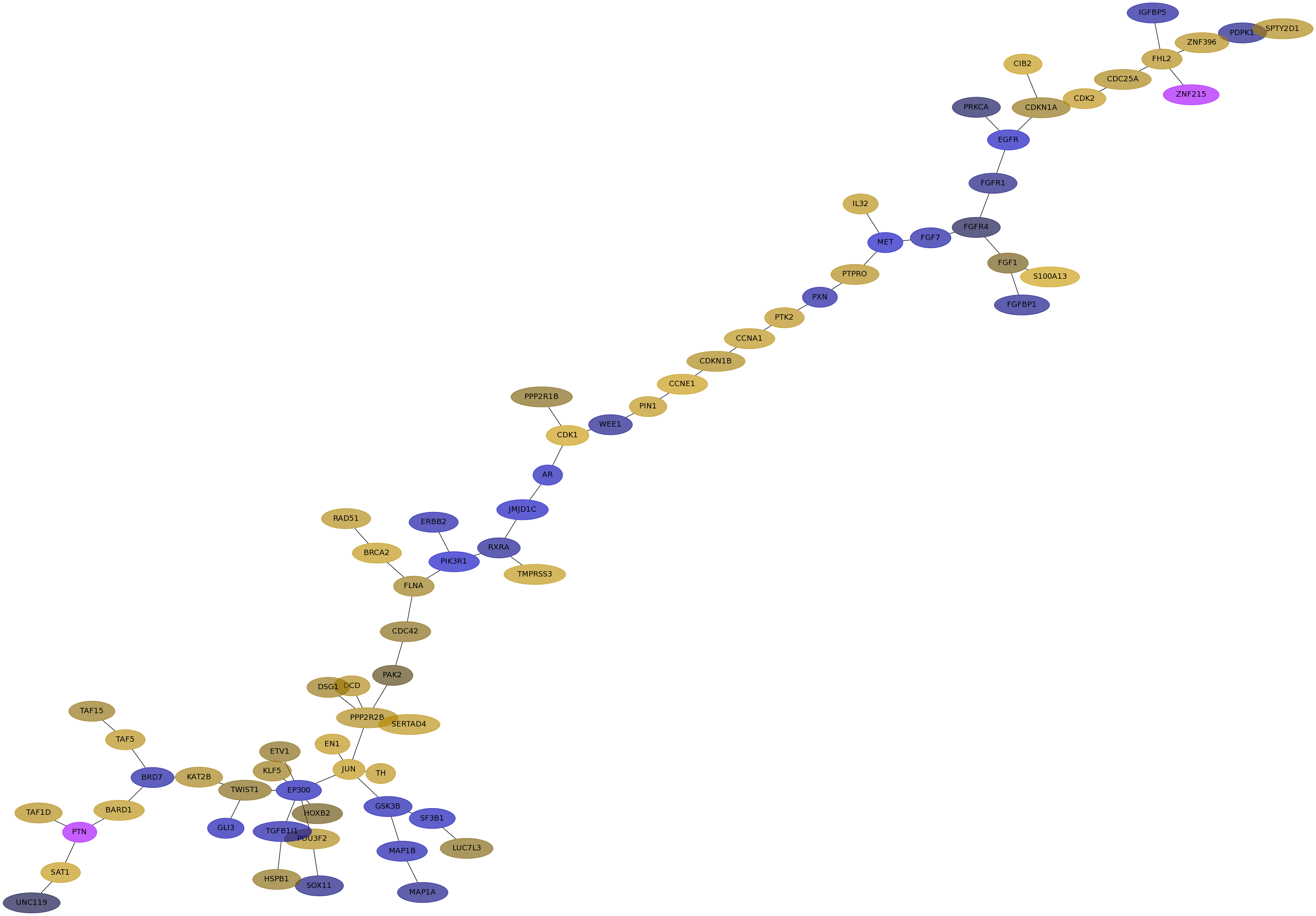
Score for each gene in subnetwork 6262 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PTPRO |   | 2 | 743 | 1988 | 1970 | 0.133 | -0.086 | -0.058 | undef | 0.134 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| CCNE1 |   | 4 | 440 | 1083 | 1067 | 0.245 | -0.141 | -0.077 | undef | -0.018 |
|---|
| PPP2R1B |   | 6 | 301 | 141 | 152 | 0.042 | 0.002 | -0.002 | undef | 0.112 |
|---|
| WEE1 |   | 4 | 440 | 658 | 649 | -0.051 | 0.163 | 0.034 | undef | 0.215 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| DCD |   | 1 | 1195 | 1988 | 1993 | 0.135 | undef | undef | undef | undef |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| CDKN1B |   | 1 | 1195 | 1988 | 1993 | 0.120 | -0.171 | 0.088 | -0.115 | -0.005 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| MET |   | 6 | 301 | 1047 | 1020 | -0.219 | 0.086 | 0.183 | -0.025 | 0.170 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| S100A13 |   | 1 | 1195 | 1988 | 1993 | 0.286 | 0.092 | 0.019 | 0.025 | 0.045 |
|---|
| SOX11 |   | 5 | 360 | 1396 | 1378 | -0.039 | -0.073 | -0.028 | 0.274 | -0.104 |
|---|
| TAF5 |   | 5 | 360 | 478 | 476 | 0.166 | -0.131 | 0.125 | undef | 0.076 |
|---|
| SERTAD4 |   | 8 | 222 | 1268 | 1225 | 0.173 | 0.169 | undef | -0.050 | undef |
|---|
| SAT1 |   | 6 | 301 | 236 | 239 | 0.219 | 0.010 | -0.066 | -0.119 | 0.063 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| ZNF396 |   | 2 | 743 | 882 | 892 | 0.151 | undef | undef | undef | undef |
|---|
| FGF1 |   | 4 | 440 | 1604 | 1572 | 0.027 | undef | undef | 0.090 | undef |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| TMPRSS3 |   | 1 | 1195 | 1988 | 1993 | 0.207 | 0.026 | -0.172 | undef | -0.132 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| TH |   | 2 | 743 | 1988 | 1970 | 0.182 | -0.018 | 0.068 | undef | 0.039 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| IL32 |   | 1 | 1195 | 1988 | 1993 | 0.170 | -0.229 | -0.100 | -0.187 | 0.039 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| ZNF215 |   | 1 | 1195 | 1988 | 1993 | undef | -0.035 | 0.194 | -0.036 | 0.163 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| PDPK1 |   | 1 | 1195 | 1988 | 1993 | -0.050 | -0.200 | -0.024 | 0.025 | 0.134 |
|---|
| SPTY2D1 |   | 1 | 1195 | 1988 | 1993 | 0.129 | 0.017 | undef | undef | undef |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| CDC25A |   | 8 | 222 | 570 | 555 | 0.113 | -0.170 | 0.151 | -0.301 | -0.003 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| PTK2 |   | 4 | 440 | 1437 | 1414 | 0.166 | -0.056 | 0.160 | -0.032 | -0.020 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TAF15 |   | 2 | 743 | 457 | 481 | 0.066 | 0.089 | 0.207 | 0.117 | 0.095 |
|---|
| FGFBP1 |   | 1 | 1195 | 1988 | 1993 | -0.051 | 0.030 | -0.018 | -0.150 | -0.086 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
GO Enrichment output for subnetwork 6262 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.434E-09 | 5.946E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.127E-09 | 5.041E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 8.404E-09 | 6.844E-06 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 7.126E-07 | 4.352E-04 |
|---|
| signal complex assembly | GO:0007172 |  | 1.674E-06 | 8.177E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.775E-06 | 1.13E-03 |
|---|
| nuclear migration | GO:0007097 |  | 2.833E-06 | 9.887E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.833E-06 | 8.651E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.365E-06 | 9.134E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.935E-06 | 9.613E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.935E-06 | 8.739E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.952E-09 | 7.101E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.003E-09 | 6.019E-06 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 8.131E-07 | 6.521E-04 |
|---|
| signal complex assembly | GO:0007172 |  | 1.909E-06 | 1.148E-03 |
|---|
| male meiosis | GO:0007140 |  | 3.133E-06 | 1.508E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.133E-06 | 1.256E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.133E-06 | 1.077E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.255E-06 | 9.791E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.837E-06 | 1.026E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.614E-06 | 1.11E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.154E-06 | 1.127E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.135E-08 | 2.611E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.968E-08 | 2.264E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.749E-06 | 1.341E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 2.08E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 3.617E-06 | 1.664E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 1.387E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.902E-06 | 1.611E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 5.694E-06 | 1.637E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.694E-06 | 1.455E-03 |
|---|
| male meiosis | GO:0007140 |  | 7.197E-06 | 1.655E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.197E-06 | 1.505E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.221E-07 | 5.935E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.839E-07 | 3.538E-04 |
|---|
| anterior/posterior pattern formation | GO:0009952 |  | 4.652E-07 | 2.858E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 4.85E-07 | 2.235E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.733E-07 | 2.113E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.003E-06 | 3.079E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.149E-06 | 3.026E-04 |
|---|
| cell fate specification | GO:0001708 |  | 2.071E-06 | 4.771E-04 |
|---|
| lung development | GO:0030324 |  | 2.504E-06 | 5.127E-04 |
|---|
| regionalization | GO:0003002 |  | 3.222E-06 | 5.938E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 3.277E-06 | 5.491E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.135E-08 | 2.611E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.968E-08 | 2.264E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.749E-06 | 1.341E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 2.08E-03 |
|---|
| cranial nerve morphogenesis | GO:0021602 |  | 3.617E-06 | 1.664E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 1.387E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.902E-06 | 1.611E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 5.694E-06 | 1.637E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.694E-06 | 1.455E-03 |
|---|
| male meiosis | GO:0007140 |  | 7.197E-06 | 1.655E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.197E-06 | 1.505E-03 |
|---|



