Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6261
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7593 | 3.188e-03 | 5.040e-04 | 7.561e-01 | 1.215e-06 |
|---|
| Loi | 0.2289 | 8.248e-02 | 1.195e-02 | 4.336e-01 | 4.273e-04 |
|---|
| Schmidt | 0.6712 | 0.000e+00 | 0.000e+00 | 4.736e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.477e-03 | 0.000e+00 |
|---|
| Wang | 0.2596 | 2.962e-03 | 4.635e-02 | 4.079e-01 | 5.600e-05 |
|---|
Expression data for subnetwork 6261 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
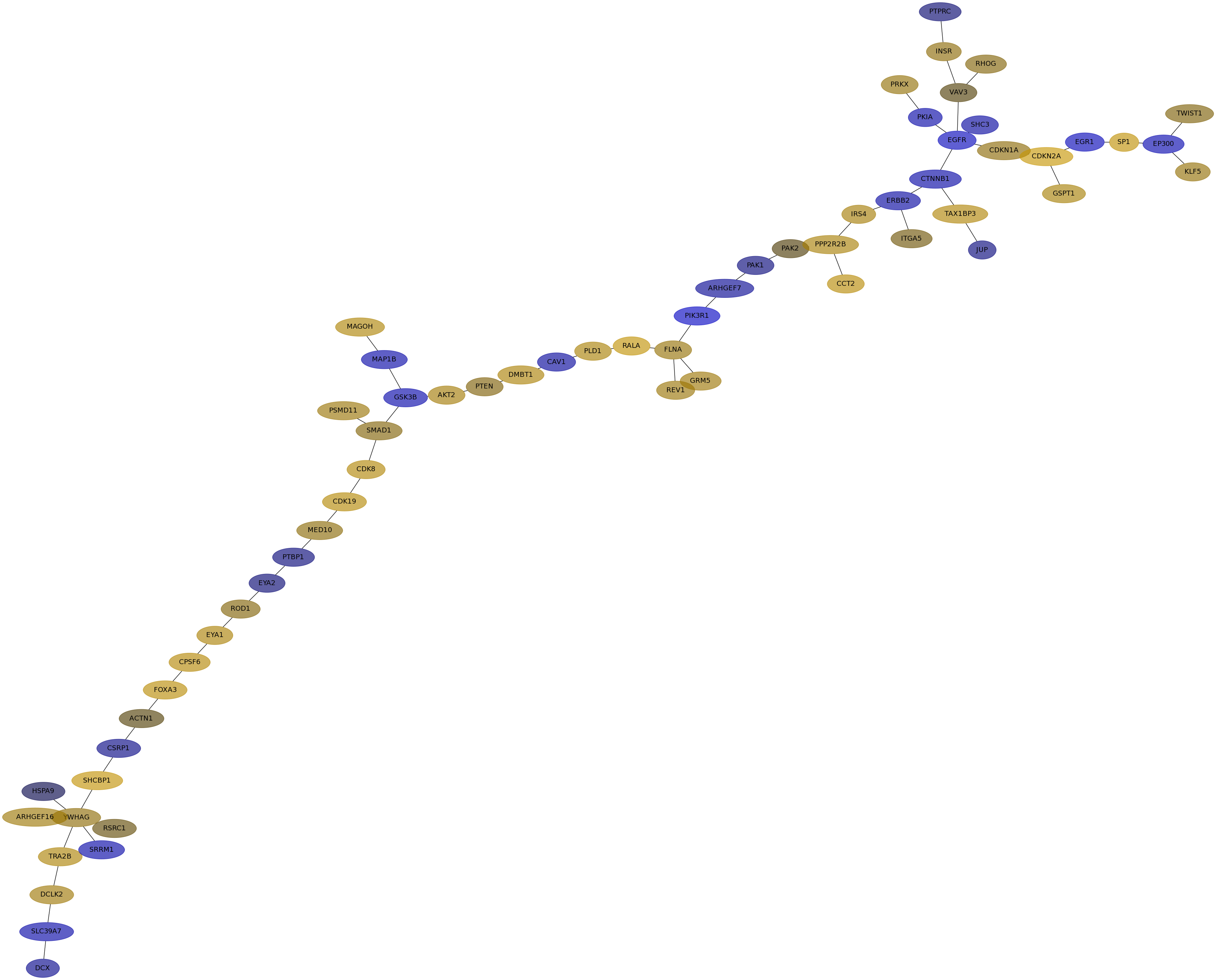
Score for each gene in subnetwork 6261 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| RHOG |   | 1 | 1195 | 1975 | 1982 | 0.049 | -0.200 | -0.109 | -0.144 | 0.151 |
|---|
| JUP |   | 1 | 1195 | 1975 | 1982 | -0.042 | 0.182 | -0.155 | 0.059 | -0.034 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| EYA1 |   | 8 | 222 | 1010 | 981 | 0.135 | -0.065 | 0.162 | 0.068 | -0.088 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| DCX |   | 1 | 1195 | 1975 | 1982 | -0.064 | -0.027 | 0.005 | 0.081 | 0.001 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| DMBT1 |   | 1 | 1195 | 1975 | 1982 | 0.134 | 0.012 | -0.065 | undef | -0.132 |
|---|
| MED10 |   | 3 | 557 | 1165 | 1151 | 0.062 | -0.149 | undef | undef | undef |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| SHC3 |   | 3 | 557 | 1165 | 1151 | -0.101 | -0.039 | -0.050 | -0.040 | -0.009 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| SLC39A7 |   | 2 | 743 | 1845 | 1831 | -0.119 | 0.157 | -0.147 | 0.185 | 0.175 |
|---|
| TRA2B |   | 1 | 1195 | 1975 | 1982 | 0.139 | -0.012 | 0.155 | 0.040 | 0.128 |
|---|
| TAX1BP3 |   | 2 | 743 | 1274 | 1260 | 0.150 | 0.219 | 0.070 | undef | 0.206 |
|---|
| PSMD11 |   | 6 | 301 | 1437 | 1408 | 0.088 | -0.090 | -0.137 | 0.060 | -0.078 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| CPSF6 |   | 6 | 301 | 318 | 330 | 0.164 | 0.052 | 0.091 | 0.129 | -0.088 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PAK1 |   | 3 | 557 | 682 | 685 | -0.040 | -0.122 | -0.143 | 0.075 | 0.006 |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| CCT2 |   | 3 | 557 | 1474 | 1450 | 0.182 | 0.008 | 0.043 | 0.055 | 0.004 |
|---|
| CSRP1 |   | 2 | 743 | 1975 | 1953 | -0.053 | 0.224 | 0.088 | 0.049 | 0.080 |
|---|
| CDK8 |   | 1 | 1195 | 1975 | 1982 | 0.157 | -0.060 | 0.111 | -0.187 | 0.065 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PTBP1 |   | 3 | 557 | 1165 | 1151 | -0.038 | -0.090 | -0.113 | 0.054 | -0.110 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| MAGOH |   | 2 | 743 | 1346 | 1333 | 0.144 | -0.199 | 0.089 | -0.052 | 0.228 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| PRKX |   | 1 | 1195 | 1975 | 1982 | 0.074 | -0.053 | -0.013 | -0.021 | -0.096 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CAV1 |   | 2 | 743 | 1510 | 1502 | -0.092 | 0.124 | 0.079 | undef | 0.119 |
|---|
| SHCBP1 |   | 1 | 1195 | 1975 | 1982 | 0.233 | -0.004 | -0.063 | undef | 0.202 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| DCLK2 |   | 1 | 1195 | 1975 | 1982 | 0.101 | 0.049 | 0.141 | undef | 0.018 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| RALA |   | 10 | 167 | 318 | 323 | 0.224 | 0.207 | -0.047 | 0.169 | 0.146 |
|---|
| HSPA9 |   | 1 | 1195 | 1975 | 1982 | -0.013 | -0.111 | 0.015 | -0.046 | 0.097 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CDK19 |   | 1 | 1195 | 1975 | 1982 | 0.171 | -0.043 | 0.090 | undef | 0.054 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ROD1 |   | 4 | 440 | 1564 | 1542 | 0.049 | 0.088 | 0.052 | 0.011 | -0.223 |
|---|
| GSPT1 |   | 2 | 743 | 1975 | 1953 | 0.123 | -0.024 | 0.036 | 0.037 | -0.040 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| SMAD1 |   | 5 | 360 | 682 | 671 | 0.049 | 0.159 | -0.062 | 0.140 | 0.048 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
GO Enrichment output for subnetwork 6261 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 9.445E-10 | 2.307E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.254E-09 | 1.531E-06 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.41E-09 | 1.148E-06 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.215E-09 | 1.353E-06 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.286E-09 | 1.117E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 6.832E-09 | 2.782E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.271E-09 | 3.236E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.588E-08 | 4.85E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 2.311E-08 | 6.274E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.165E-08 | 7.732E-06 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 6.574E-08 | 1.46E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.293E-09 | 3.11E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.596E-09 | 1.92E-06 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.801E-09 | 1.445E-06 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.868E-09 | 1.725E-06 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.944E-09 | 1.417E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 8.799E-09 | 3.528E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.417E-08 | 4.871E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.045E-08 | 6.15E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 2.825E-08 | 7.553E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.074E-08 | 9.803E-06 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 8.971E-08 | 1.962E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 6.995E-08 | 1.609E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 7.914E-08 | 9.101E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.13E-07 | 8.667E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.139E-07 | 1.805E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 3.139E-07 | 1.444E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.807E-07 | 1.459E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.162E-07 | 2.682E-04 |
|---|
| regulation of protein stability | GO:0031647 |  | 1.655E-06 | 4.759E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.468E-06 | 6.308E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.914E-06 | 1.13E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.914E-06 | 1.027E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.411E-08 | 2.601E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.754E-07 | 4.381E-04 |
|---|
| receptor clustering | GO:0043113 |  | 2.255E-06 | 1.385E-03 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.255E-06 | 1.039E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.822E-06 | 1.04E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 2.855E-06 | 8.769E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.235E-06 | 8.516E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.201E-06 | 9.679E-04 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.347E-05 | 2.757E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.521E-05 | 2.804E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.538E-05 | 2.577E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 6.995E-08 | 1.609E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 7.914E-08 | 9.101E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.13E-07 | 8.667E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.139E-07 | 1.805E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 3.139E-07 | 1.444E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.807E-07 | 1.459E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.162E-07 | 2.682E-04 |
|---|
| regulation of protein stability | GO:0031647 |  | 1.655E-06 | 4.759E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.468E-06 | 6.308E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.914E-06 | 1.13E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.914E-06 | 1.027E-03 |
|---|



