Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6241
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7596 | 3.170e-03 | 5.010e-04 | 7.552e-01 | 1.199e-06 |
|---|
| Loi | 0.2288 | 8.265e-02 | 1.200e-02 | 4.340e-01 | 4.303e-04 |
|---|
| Schmidt | 0.6713 | 0.000e+00 | 0.000e+00 | 4.728e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.484e-03 | 0.000e+00 |
|---|
| Wang | 0.2594 | 2.981e-03 | 4.652e-02 | 4.087e-01 | 5.667e-05 |
|---|
Expression data for subnetwork 6241 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
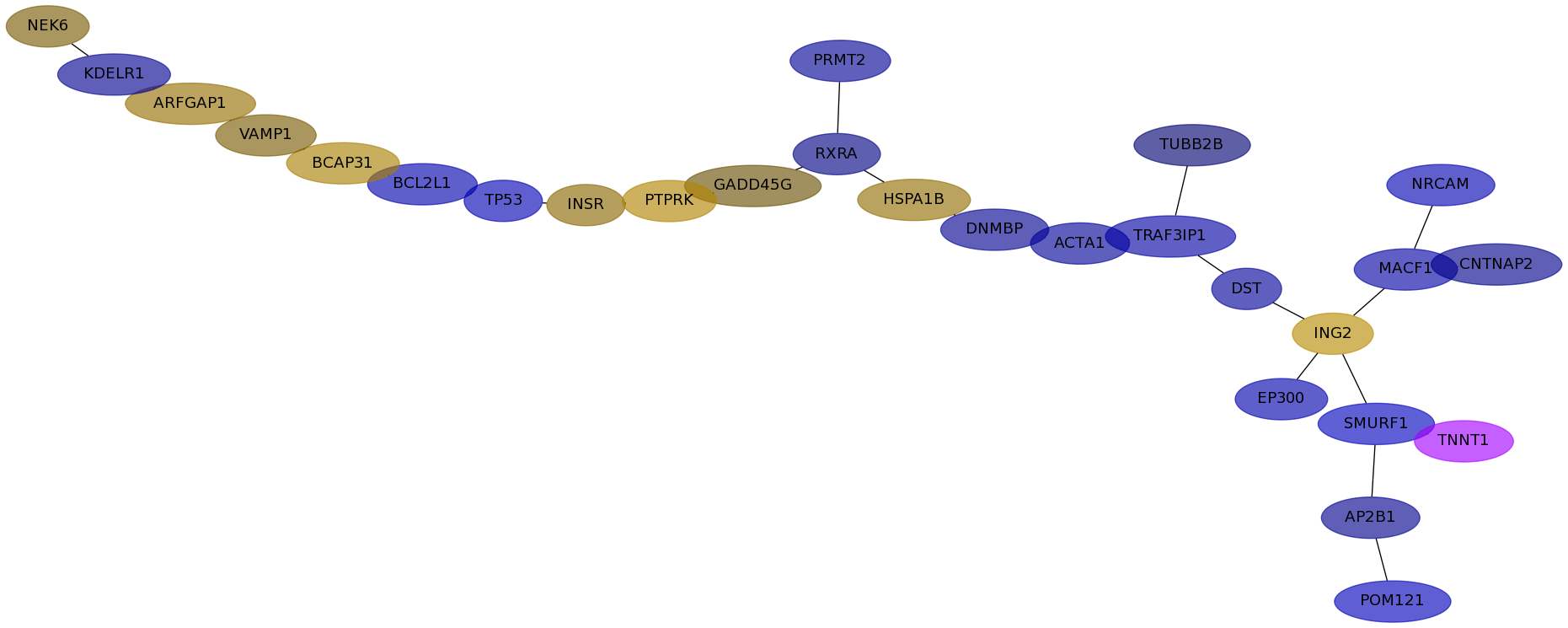
Subnetwork structure for each dataset
- IPC
Score for each gene in subnetwork 6241 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| DNMBP |   | 1 | 1195 | 1914 | 1926 | -0.079 | 0.152 | 0.048 | 0.083 | 0.035 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| ING2 |   | 3 | 557 | 1 | 63 | 0.189 | 0.168 | 0.051 | 0.026 | -0.054 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CNTNAP2 |   | 1 | 1195 | 1914 | 1926 | -0.063 | 0.156 | -0.119 | 0.213 | -0.055 |
|---|
| BCL2L1 |   | 1 | 1195 | 1914 | 1926 | -0.149 | 0.082 | -0.191 | 0.044 | -0.115 |
|---|
| PRMT2 |   | 5 | 360 | 957 | 938 | -0.081 | 0.046 | 0.089 | 0.235 | -0.128 |
|---|
| MACF1 |   | 1 | 1195 | 1914 | 1926 | -0.122 | 0.265 | -0.014 | -0.035 | -0.011 |
|---|
| NRCAM |   | 2 | 743 | 1842 | 1829 | -0.163 | 0.111 | 0.112 | 0.090 | -0.138 |
|---|
| BCAP31 |   | 2 | 743 | 1803 | 1782 | 0.131 | 0.020 | 0.055 | 0.168 | -0.018 |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| HSPA1B |   | 2 | 743 | 1914 | 1897 | 0.075 | 0.117 | -0.061 | undef | 0.223 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| ARFGAP1 |   | 1 | 1195 | 1914 | 1926 | 0.082 | 0.167 | 0.038 | undef | 0.212 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| VAMP1 |   | 1 | 1195 | 1914 | 1926 | 0.042 | -0.029 | -0.197 | -0.088 | 0.013 |
|---|
| NEK6 |   | 3 | 557 | 842 | 834 | 0.041 | -0.049 | undef | 0.271 | undef |
|---|
| TNNT1 |   | 3 | 557 | 1914 | 1877 | undef | undef | undef | 0.145 | undef |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| ACTA1 |   | 4 | 440 | 83 | 112 | -0.090 | -0.041 | 0.145 | 0.248 | 0.132 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| AP2B1 |   | 6 | 301 | 780 | 764 | -0.070 | -0.106 | 0.070 | 0.252 | 0.094 |
|---|
| SMURF1 |   | 5 | 360 | 682 | 671 | -0.222 | 0.236 | -0.049 | undef | -0.006 |
|---|
| KDELR1 |   | 2 | 743 | 842 | 854 | -0.073 | -0.029 | -0.123 | 0.288 | 0.004 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
GO Enrichment output for subnetwork 6241 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.136E-07 | 2.776E-04 |
|---|
| protein targeting to ER | GO:0045047 |  | 9.483E-07 | 1.158E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.075E-06 | 3.319E-03 |
|---|
| neuron recognition | GO:0008038 |  | 6.249E-06 | 3.817E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.264E-05 | 6.175E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 2.529E-05 | 0.0102971 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 3.206E-05 | 0.01118763 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.106E-05 | 0.01559396 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 5.106E-05 | 0.0138613 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 5.106E-05 | 0.01247517 |
|---|
| muscle cell proliferation | GO:0033002 |  | 5.106E-05 | 0.01134106 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.361E-07 | 3.276E-04 |
|---|
| protein targeting to ER | GO:0045047 |  | 1.136E-06 | 1.366E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.582E-06 | 3.675E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.879E-06 | 2.934E-03 |
|---|
| neuron recognition | GO:0008038 |  | 7.479E-06 | 3.599E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.512E-05 | 6.063E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 2.666E-05 | 9.163E-03 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 3.832E-05 | 0.01152617 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.768E-05 | 0.01541879 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 5.768E-05 | 0.01387691 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 5.768E-05 | 0.01261538 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.329E-07 | 5.358E-04 |
|---|
| protein targeting to ER | GO:0045047 |  | 1.941E-06 | 2.232E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 5.036E-06 | 3.861E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 8.323E-06 | 4.786E-03 |
|---|
| neuron recognition | GO:0008038 |  | 1.038E-05 | 4.776E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.851E-05 | 7.094E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 3.464E-05 | 0.01138124 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 4.534E-05 | 0.01303466 |
|---|
| mesoderm formation | GO:0001707 |  | 7.278E-05 | 0.01859972 |
|---|
| protein targeting to membrane | GO:0006612 |  | 8.101E-05 | 0.0186329 |
|---|
| formation of primary germ layer | GO:0001704 |  | 8.101E-05 | 0.016939 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein tetramerization | GO:0051262 |  | 5.899E-06 | 0.01087208 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 9.35E-06 | 8.616E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.012E-05 | 6.217E-03 |
|---|
| striated muscle tissue development | GO:0014706 |  | 3.54E-05 | 0.0163098 |
|---|
| muscle tissue development | GO:0060537 |  | 4.211E-05 | 0.01552113 |
|---|
| regulation of gene-specific transcription | GO:0032583 |  | 4.64E-05 | 0.01425126 |
|---|
| heart morphogenesis | GO:0003007 |  | 4.64E-05 | 0.01221536 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 7.987E-05 | 0.01840053 |
|---|
| negative regulation of neuroblast proliferation | GO:0007406 |  | 7.987E-05 | 0.01635603 |
|---|
| musculoskeletal movement | GO:0050881 |  | 7.987E-05 | 0.01472042 |
|---|
| cardiac cell differentiation | GO:0035051 |  | 7.987E-05 | 0.0133822 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.329E-07 | 5.358E-04 |
|---|
| protein targeting to ER | GO:0045047 |  | 1.941E-06 | 2.232E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 5.036E-06 | 3.861E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 8.323E-06 | 4.786E-03 |
|---|
| neuron recognition | GO:0008038 |  | 1.038E-05 | 4.776E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.851E-05 | 7.094E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 3.464E-05 | 0.01138124 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 4.534E-05 | 0.01303466 |
|---|
| mesoderm formation | GO:0001707 |  | 7.278E-05 | 0.01859972 |
|---|
| protein targeting to membrane | GO:0006612 |  | 8.101E-05 | 0.0186329 |
|---|
| formation of primary germ layer | GO:0001704 |  | 8.101E-05 | 0.016939 |
|---|



