Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6240
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7596 | 3.168e-03 | 5.000e-04 | 7.551e-01 | 1.196e-06 |
|---|
| Loi | 0.2287 | 8.271e-02 | 1.201e-02 | 4.341e-01 | 4.314e-04 |
|---|
| Schmidt | 0.6713 | 0.000e+00 | 0.000e+00 | 4.727e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.485e-03 | 0.000e+00 |
|---|
| Wang | 0.2594 | 2.980e-03 | 4.651e-02 | 4.086e-01 | 5.663e-05 |
|---|
Expression data for subnetwork 6240 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
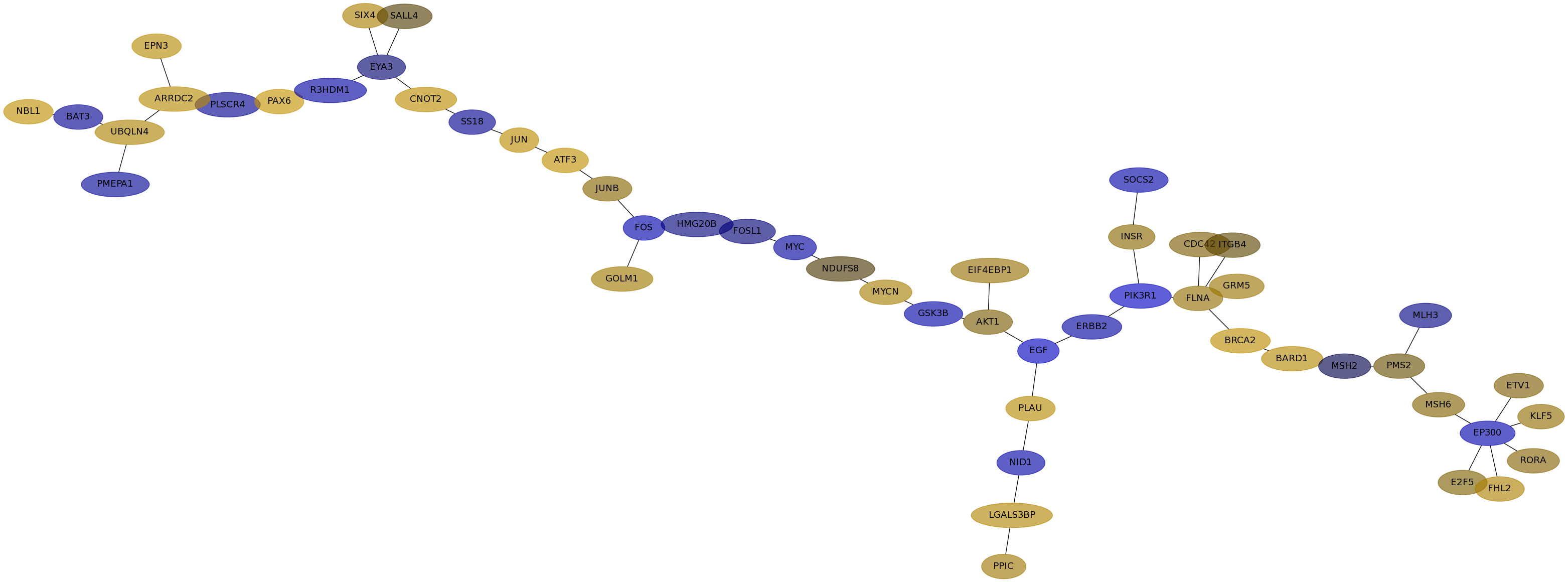
Score for each gene in subnetwork 6240 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NBL1 |   | 1 | 1195 | 1893 | 1903 | 0.230 | -0.068 | undef | undef | undef |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| E2F5 |   | 2 | 743 | 1274 | 1260 | 0.048 | -0.087 | 0.002 | -0.032 | -0.074 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| SOCS2 |   | 3 | 557 | 1688 | 1660 | -0.130 | -0.028 | 0.051 | -0.171 | 0.027 |
|---|
| PMS2 |   | 2 | 743 | 1425 | 1421 | 0.029 | 0.095 | -0.017 | -0.040 | -0.026 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RORA |   | 2 | 743 | 478 | 490 | 0.056 | 0.272 | 0.051 | undef | 0.062 |
|---|
| MLH3 |   | 2 | 743 | 1425 | 1421 | -0.051 | 0.158 | -0.030 | -0.112 | 0.103 |
|---|
| SIX4 |   | 2 | 743 | 1893 | 1869 | 0.135 | -0.094 | undef | -0.001 | undef |
|---|
| LGALS3BP |   | 1 | 1195 | 1893 | 1903 | 0.167 | 0.012 | -0.002 | 0.101 | -0.197 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| GOLM1 |   | 8 | 222 | 1534 | 1489 | 0.109 | 0.161 | -0.217 | 0.170 | -0.060 |
|---|
| SS18 |   | 1 | 1195 | 1893 | 1903 | -0.071 | 0.040 | -0.101 | 0.047 | -0.085 |
|---|
| PPIC |   | 2 | 743 | 1461 | 1451 | 0.103 | 0.192 | -0.225 | 0.151 | 0.024 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| MSH2 |   | 1 | 1195 | 1893 | 1903 | -0.013 | -0.128 | 0.055 | -0.030 | -0.068 |
|---|
| NDUFS8 |   | 3 | 557 | 696 | 695 | 0.013 | -0.088 | -0.150 | 0.176 | 0.030 |
|---|
| MYCN |   | 7 | 256 | 764 | 756 | 0.125 | 0.057 | -0.003 | -0.081 | -0.084 |
|---|
| ARRDC2 |   | 4 | 440 | 696 | 689 | 0.192 | -0.100 | undef | undef | undef |
|---|
| BAT3 |   | 4 | 440 | 236 | 262 | -0.078 | 0.072 | -0.057 | 0.110 | 0.069 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| UBQLN4 |   | 4 | 440 | 696 | 689 | 0.143 | -0.114 | undef | -0.213 | undef |
|---|
| NID1 |   | 1 | 1195 | 1893 | 1903 | -0.117 | 0.170 | 0.059 | 0.184 | 0.166 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| SALL4 |   | 1 | 1195 | 1893 | 1903 | 0.017 | 0.112 | undef | 0.093 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| HMG20B |   | 2 | 743 | 1893 | 1869 | -0.045 | 0.146 | -0.151 | -0.074 | -0.155 |
|---|
| R3HDM1 |   | 4 | 440 | 318 | 337 | -0.108 | -0.192 | -0.049 | 0.082 | 0.223 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| CNOT2 |   | 1 | 1195 | 1893 | 1903 | 0.202 | 0.114 | 0.112 | -0.007 | 0.123 |
|---|
| EPN3 |   | 1 | 1195 | 1893 | 1903 | 0.185 | 0.179 | 0.001 | 0.132 | 0.081 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| PMEPA1 |   | 3 | 557 | 1294 | 1276 | -0.085 | 0.137 | 0.167 | 0.087 | 0.102 |
|---|
| EIF4EBP1 |   | 3 | 557 | 1222 | 1210 | 0.086 | -0.201 | -0.147 | 0.183 | 0.216 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| FOSL1 |   | 11 | 148 | 366 | 360 | -0.040 | 0.164 | 0.010 | 0.128 | 0.005 |
|---|
GO Enrichment output for subnetwork 6240 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 9.77E-09 | 2.387E-05 |
|---|
| mismatch repair | GO:0006298 |  | 2.843E-07 | 3.472E-04 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 4.123E-07 | 3.357E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 4.123E-07 | 2.518E-04 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 5.251E-07 | 2.566E-04 |
|---|
| nuclear migration | GO:0007097 |  | 8.225E-07 | 3.349E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.225E-07 | 2.87E-04 |
|---|
| determination of adult lifespan | GO:0008340 |  | 8.225E-07 | 2.512E-04 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 9.83E-07 | 2.668E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.217E-06 | 2.972E-04 |
|---|
| somatic diversification of immune receptors via germline recombination within a single locus | GO:0002562 |  | 1.42E-06 | 3.154E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.347E-08 | 3.241E-05 |
|---|
| mismatch repair | GO:0006298 |  | 1.813E-07 | 2.181E-04 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 5.253E-07 | 4.213E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.253E-07 | 3.16E-04 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 7.223E-07 | 3.476E-04 |
|---|
| male meiosis | GO:0007140 |  | 1.048E-06 | 4.201E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.048E-06 | 3.601E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.048E-06 | 3.151E-04 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.048E-06 | 2.801E-04 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 1.351E-06 | 3.251E-04 |
|---|
| response to UV | GO:0009411 |  | 1.766E-06 | 3.862E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.717E-08 | 3.949E-05 |
|---|
| mismatch repair | GO:0006298 |  | 2.463E-07 | 2.832E-04 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 8.638E-07 | 6.622E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.063E-06 | 6.111E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.063E-06 | 4.889E-04 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.063E-06 | 4.074E-04 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 1.819E-06 | 5.978E-04 |
|---|
| regulation of helicase activity | GO:0051095 |  | 2.118E-06 | 6.09E-04 |
|---|
| male meiosis | GO:0007140 |  | 2.118E-06 | 5.413E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.118E-06 | 4.872E-04 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 2.118E-06 | 4.429E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| muscle cell migration | GO:0014812 |  | 9.085E-07 | 1.674E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.811E-06 | 1.669E-03 |
|---|
| multicellular organismal aging | GO:0010259 |  | 1.811E-06 | 1.113E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.554E-06 | 2.098E-03 |
|---|
| immunoglobulin production during immune response | GO:0002381 |  | 5.039E-06 | 1.857E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 6.277E-06 | 1.928E-03 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 7.535E-06 | 1.984E-03 |
|---|
| mismatch repair | GO:0006298 |  | 1.073E-05 | 2.472E-03 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.111E-05 | 2.275E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.471E-05 | 2.711E-03 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 1.471E-05 | 2.464E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.717E-08 | 3.949E-05 |
|---|
| mismatch repair | GO:0006298 |  | 2.463E-07 | 2.832E-04 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 8.638E-07 | 6.622E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.063E-06 | 6.111E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.063E-06 | 4.889E-04 |
|---|
| determination of adult lifespan | GO:0008340 |  | 1.063E-06 | 4.074E-04 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 1.819E-06 | 5.978E-04 |
|---|
| regulation of helicase activity | GO:0051095 |  | 2.118E-06 | 6.09E-04 |
|---|
| male meiosis | GO:0007140 |  | 2.118E-06 | 5.413E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.118E-06 | 4.872E-04 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 2.118E-06 | 4.429E-04 |
|---|



