Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6231
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7598 | 3.160e-03 | 4.990e-04 | 7.547e-01 | 1.190e-06 |
|---|
| Loi | 0.2284 | 8.322e-02 | 1.215e-02 | 4.353e-01 | 4.402e-04 |
|---|
| Schmidt | 0.6715 | 0.000e+00 | 0.000e+00 | 4.697e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.486e-03 | 0.000e+00 |
|---|
| Wang | 0.2594 | 2.992e-03 | 4.661e-02 | 4.091e-01 | 5.705e-05 |
|---|
Expression data for subnetwork 6231 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
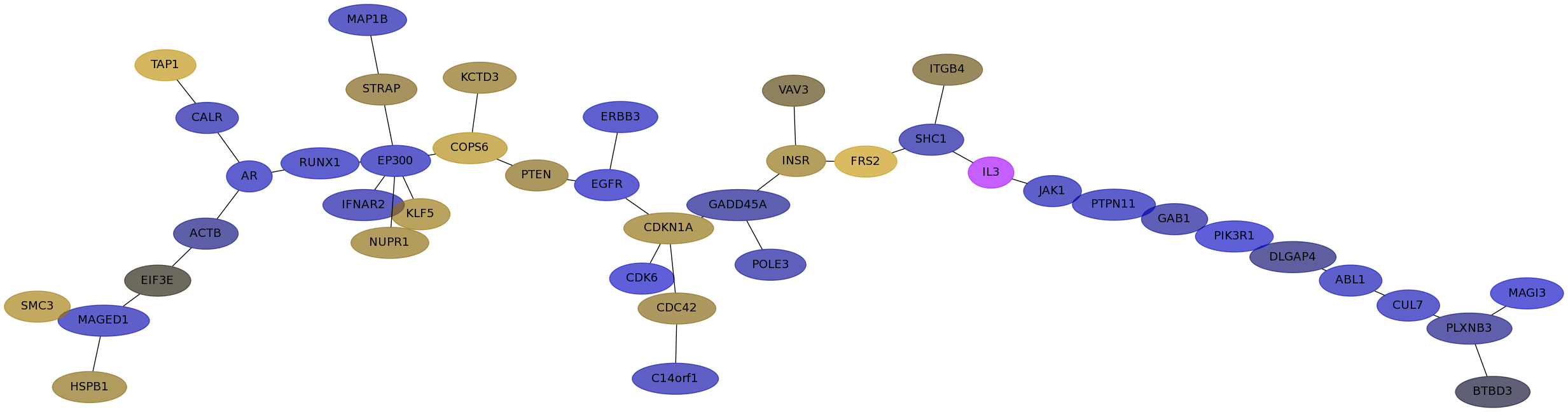
Score for each gene in subnetwork 6231 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PLXNB3 |   | 1 | 1195 | 1922 | 1935 | -0.043 | 0.152 | -0.113 | 0.166 | -0.070 |
|---|
| IFNAR2 |   | 4 | 440 | 1558 | 1528 | -0.100 | 0.055 | 0.106 | -0.294 | -0.058 |
|---|
| CUL7 |   | 3 | 557 | 1621 | 1593 | -0.157 | 0.176 | -0.108 | -0.006 | 0.104 |
|---|
| MAGED1 |   | 5 | 360 | 842 | 828 | -0.141 | 0.284 | -0.152 | 0.240 | 0.045 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| TAP1 |   | 2 | 743 | 1922 | 1902 | 0.210 | -0.116 | -0.118 | -0.078 | -0.146 |
|---|
| EIF3E |   | 1 | 1195 | 1922 | 1935 | 0.003 | -0.120 | 0.137 | -0.048 | -0.003 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| CALR |   | 4 | 440 | 525 | 520 | -0.105 | 0.002 | -0.061 | -0.094 | -0.075 |
|---|
| SMC3 |   | 5 | 360 | 614 | 612 | 0.105 | -0.096 | 0.238 | 0.137 | 0.047 |
|---|
| C14orf1 |   | 1 | 1195 | 1922 | 1935 | -0.116 | 0.078 | -0.118 | -0.011 | 0.041 |
|---|
| SHC1 |   | 4 | 440 | 478 | 479 | -0.090 | -0.041 | -0.005 | 0.169 | -0.138 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| ERBB3 |   | 3 | 557 | 1922 | 1888 | -0.171 | -0.007 | -0.141 | -0.019 | -0.098 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| BTBD3 |   | 1 | 1195 | 1922 | 1935 | -0.005 | 0.112 | 0.152 | -0.035 | 0.056 |
|---|
| KCTD3 |   | 3 | 557 | 913 | 908 | 0.051 | 0.016 | 0.086 | 0.256 | 0.053 |
|---|
| FRS2 |   | 2 | 743 | 1083 | 1084 | 0.252 | 0.093 | 0.174 | -0.092 | -0.095 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| STRAP |   | 7 | 256 | 412 | 414 | 0.035 | -0.128 | 0.074 | 0.154 | -0.049 |
|---|
| IL3 |   | 2 | 743 | 1867 | 1849 | undef | -0.029 | 0.000 | 0.153 | undef |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAGI3 |   | 2 | 743 | 1842 | 1829 | -0.233 | 0.199 | undef | undef | undef |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| JAK1 |   | 5 | 360 | 696 | 680 | -0.140 | 0.034 | -0.176 | 0.111 | 0.159 |
|---|
| NUPR1 |   | 3 | 557 | 1222 | 1210 | 0.057 | -0.031 | -0.241 | 0.145 | 0.013 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| ACTB |   | 3 | 557 | 1274 | 1258 | -0.039 | 0.283 | -0.061 | 0.164 | 0.003 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| RUNX1 |   | 5 | 360 | 906 | 890 | -0.161 | 0.140 | 0.016 | 0.102 | 0.025 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| GAB1 |   | 5 | 360 | 1102 | 1076 | -0.069 | 0.084 | -0.117 | 0.116 | -0.212 |
|---|
GO Enrichment output for subnetwork 6231 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 9.34E-10 | 2.282E-06 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.174E-09 | 1.434E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.981E-09 | 1.613E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.794E-09 | 1.706E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.5E-09 | 3.176E-06 |
|---|
| organ growth | GO:0035265 |  | 6.5E-09 | 2.647E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 6.5E-09 | 2.269E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 6.5E-09 | 1.985E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.296E-08 | 3.519E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.296E-08 | 3.167E-06 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 2.168E-08 | 4.814E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.195E-09 | 2.874E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.995E-09 | 3.602E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.573E-09 | 2.866E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.312E-09 | 4.999E-06 |
|---|
| organ growth | GO:0035265 |  | 8.312E-09 | 4E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 8.312E-09 | 3.333E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 8.312E-09 | 2.857E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.657E-08 | 4.984E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.657E-08 | 4.43E-06 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 2.934E-08 | 7.058E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 2.974E-08 | 6.504E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.924E-09 | 4.426E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.753E-09 | 6.616E-06 |
|---|
| organ growth | GO:0035265 |  | 1.338E-08 | 1.026E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.338E-08 | 7.692E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 2.666E-08 | 1.227E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 4.141E-08 | 1.587E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.075E-07 | 3.534E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.092E-07 | 3.138E-05 |
|---|
| negative regulation of secretion | GO:0051048 |  | 2.569E-07 | 6.564E-05 |
|---|
| activation of MAPK activity | GO:0000187 |  | 2.615E-07 | 6.014E-05 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 3.563E-07 | 7.449E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of DNA replication | GO:0045740 |  | 3.445E-07 | 6.349E-04 |
|---|
| positive regulation of DNA metabolic process | GO:0051054 |  | 3.415E-06 | 3.147E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.543E-06 | 2.176E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 3.72E-06 | 1.714E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.082E-05 | 3.989E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.082E-05 | 3.324E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.424E-05 | 3.748E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 1.564E-05 | 3.602E-03 |
|---|
| striated muscle tissue development | GO:0014706 |  | 1.684E-05 | 3.448E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.771E-05 | 3.264E-03 |
|---|
| muscle tissue development | GO:0060537 |  | 2.088E-05 | 3.498E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.924E-09 | 4.426E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.753E-09 | 6.616E-06 |
|---|
| organ growth | GO:0035265 |  | 1.338E-08 | 1.026E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.338E-08 | 7.692E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 2.666E-08 | 1.227E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 4.141E-08 | 1.587E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.075E-07 | 3.534E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.092E-07 | 3.138E-05 |
|---|
| negative regulation of secretion | GO:0051048 |  | 2.569E-07 | 6.564E-05 |
|---|
| activation of MAPK activity | GO:0000187 |  | 2.615E-07 | 6.014E-05 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 3.563E-07 | 7.449E-05 |
|---|



