Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6223
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7598 | 3.159e-03 | 4.990e-04 | 7.546e-01 | 1.190e-06 |
|---|
| Loi | 0.2282 | 8.354e-02 | 1.224e-02 | 4.361e-01 | 4.458e-04 |
|---|
| Schmidt | 0.6717 | 0.000e+00 | 0.000e+00 | 4.673e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.493e-03 | 0.000e+00 |
|---|
| Wang | 0.2593 | 3.002e-03 | 4.669e-02 | 4.095e-01 | 5.740e-05 |
|---|
Expression data for subnetwork 6223 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
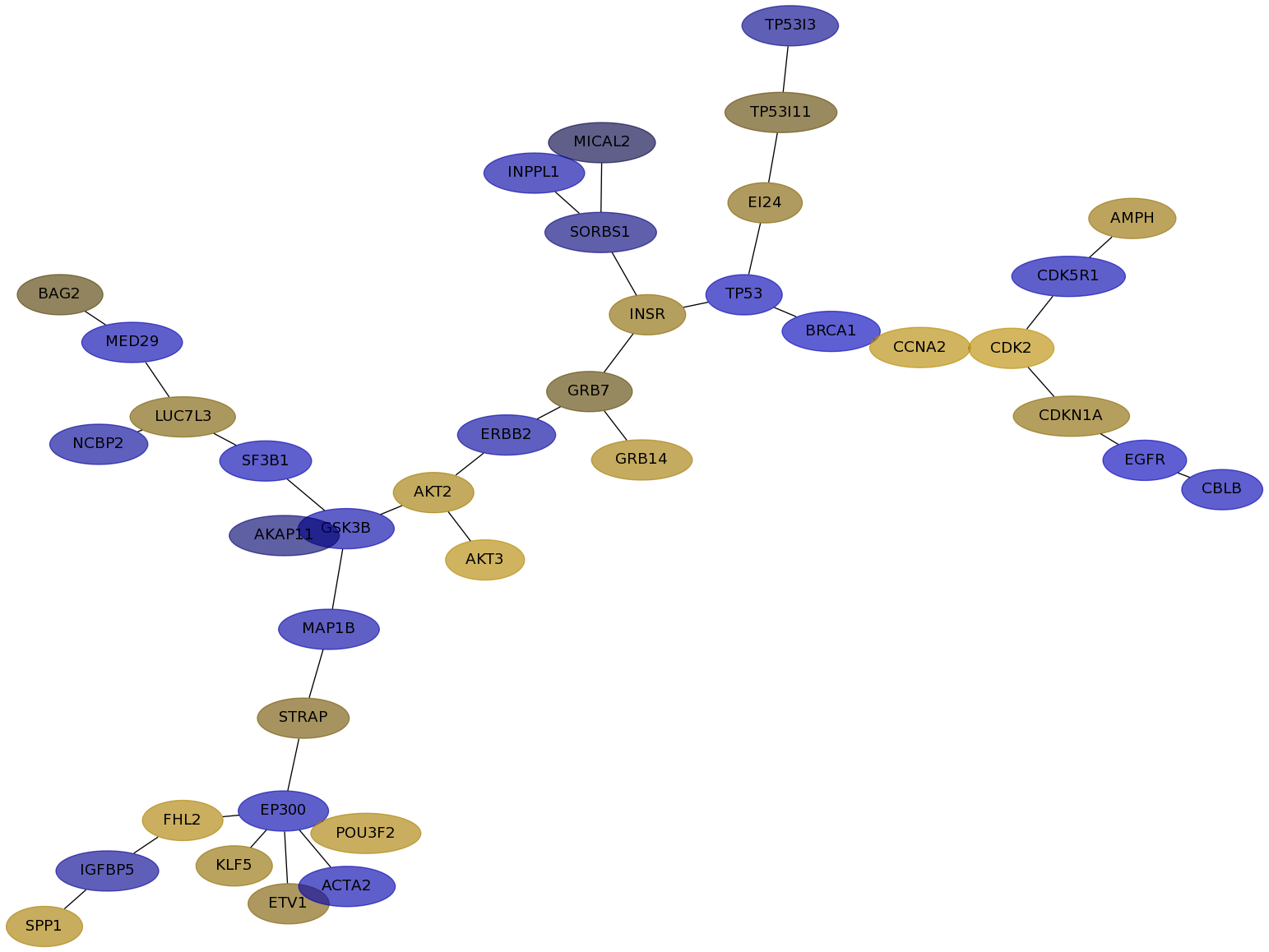
Score for each gene in subnetwork 6223 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| INPPL1 |   | 6 | 301 | 141 | 152 | -0.122 | 0.072 | -0.087 | 0.174 | 0.156 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| TP53I11 |   | 1 | 1195 | 1949 | 1955 | 0.022 | 0.058 | -0.285 | 0.335 | 0.119 |
|---|
| BAG2 |   | 4 | 440 | 1793 | 1755 | 0.016 | 0.178 | 0.152 | -0.249 | 0.083 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| AMPH |   | 7 | 256 | 83 | 102 | 0.082 | 0.000 | -0.104 | 0.024 | 0.241 |
|---|
| EI24 |   | 1 | 1195 | 1949 | 1955 | 0.051 | 0.078 | 0.107 | 0.018 | 0.070 |
|---|
| ACTA2 |   | 2 | 743 | 1949 | 1932 | -0.147 | 0.256 | 0.125 | 0.102 | 0.220 |
|---|
| MED29 |   | 1 | 1195 | 1949 | 1955 | -0.145 | -0.059 | undef | 0.114 | undef |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| AKAP11 |   | 4 | 440 | 457 | 458 | -0.034 | -0.099 | -0.038 | 0.251 | 0.017 |
|---|
| STRAP |   | 7 | 256 | 412 | 414 | 0.035 | -0.128 | 0.074 | 0.154 | -0.049 |
|---|
| MICAL2 |   | 4 | 440 | 1659 | 1629 | -0.012 | 0.143 | -0.042 | -0.145 | 0.161 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| CBLB |   | 2 | 743 | 525 | 547 | -0.168 | 0.266 | -0.248 | undef | 0.223 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| BRCA1 |   | 3 | 557 | 1534 | 1516 | -0.201 | -0.078 | -0.087 | -0.166 | 0.168 |
|---|
| TP53I3 |   | 1 | 1195 | 1949 | 1955 | -0.065 | 0.133 | 0.123 | 0.016 | -0.007 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| AKT3 |   | 5 | 360 | 1083 | 1066 | 0.172 | -0.058 | 0.087 | 0.121 | 0.080 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| NCBP2 |   | 3 | 557 | 614 | 623 | -0.087 | 0.170 | 0.311 | -0.055 | -0.052 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
GO Enrichment output for subnetwork 6223 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.78E-07 | 4.349E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 2.152E-07 | 2.629E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.76E-07 | 2.248E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 4.149E-07 | 2.534E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.198E-06 | 5.852E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.365E-06 | 3.406E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 8.789E-06 | 3.067E-03 |
|---|
| positive regulation of neuron apoptosis | GO:0043525 |  | 1.178E-05 | 3.598E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.71E-05 | 4.641E-03 |
|---|
| response to UV | GO:0009411 |  | 1.801E-05 | 4.401E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.936E-05 | 4.299E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 2.931E-07 | 7.052E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.944E-07 | 3.542E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.149E-07 | 3.328E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 6.234E-07 | 3.75E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.63E-06 | 7.845E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.313E-05 | 5.266E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.367E-05 | 4.697E-03 |
|---|
| positive regulation of neuron apoptosis | GO:0043525 |  | 1.601E-05 | 4.815E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.94E-05 | 5.185E-03 |
|---|
| response to UV | GO:0009411 |  | 2.318E-05 | 5.576E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.886E-05 | 6.313E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.044E-07 | 1.39E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 6.961E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 6.659E-07 | 5.105E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.782E-07 | 5.624E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.328E-06 | 6.108E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.614E-05 | 0.01001923 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.63E-05 | 8.64E-03 |
|---|
| positive regulation of neuron apoptosis | GO:0043525 |  | 2.941E-05 | 8.457E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.468E-05 | 8.863E-03 |
|---|
| response to UV | GO:0009411 |  | 3.787E-05 | 8.709E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.37E-05 | 9.137E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ER overload response | GO:0006983 |  | 1.484E-06 | 2.734E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.053E-06 | 1.892E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.109E-06 | 2.524E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.594E-06 | 2.577E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 8.519E-06 | 3.14E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.206E-05 | 3.704E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.206E-05 | 3.175E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.756E-05 | 4.045E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.078E-05 | 4.255E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.078E-05 | 3.83E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 2.078E-05 | 3.482E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.044E-07 | 1.39E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 6.961E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 6.659E-07 | 5.105E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.782E-07 | 5.624E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.328E-06 | 6.108E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.614E-05 | 0.01001923 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.63E-05 | 8.64E-03 |
|---|
| positive regulation of neuron apoptosis | GO:0043525 |  | 2.941E-05 | 8.457E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.468E-05 | 8.863E-03 |
|---|
| response to UV | GO:0009411 |  | 3.787E-05 | 8.709E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.37E-05 | 9.137E-03 |
|---|



