Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6222
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7599 | 3.152e-03 | 4.970e-04 | 7.543e-01 | 1.182e-06 |
|---|
| Loi | 0.2282 | 8.362e-02 | 1.226e-02 | 4.363e-01 | 4.473e-04 |
|---|
| Schmidt | 0.6718 | 0.000e+00 | 0.000e+00 | 4.667e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.494e-03 | 0.000e+00 |
|---|
| Wang | 0.2593 | 3.008e-03 | 4.675e-02 | 4.097e-01 | 5.761e-05 |
|---|
Expression data for subnetwork 6222 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
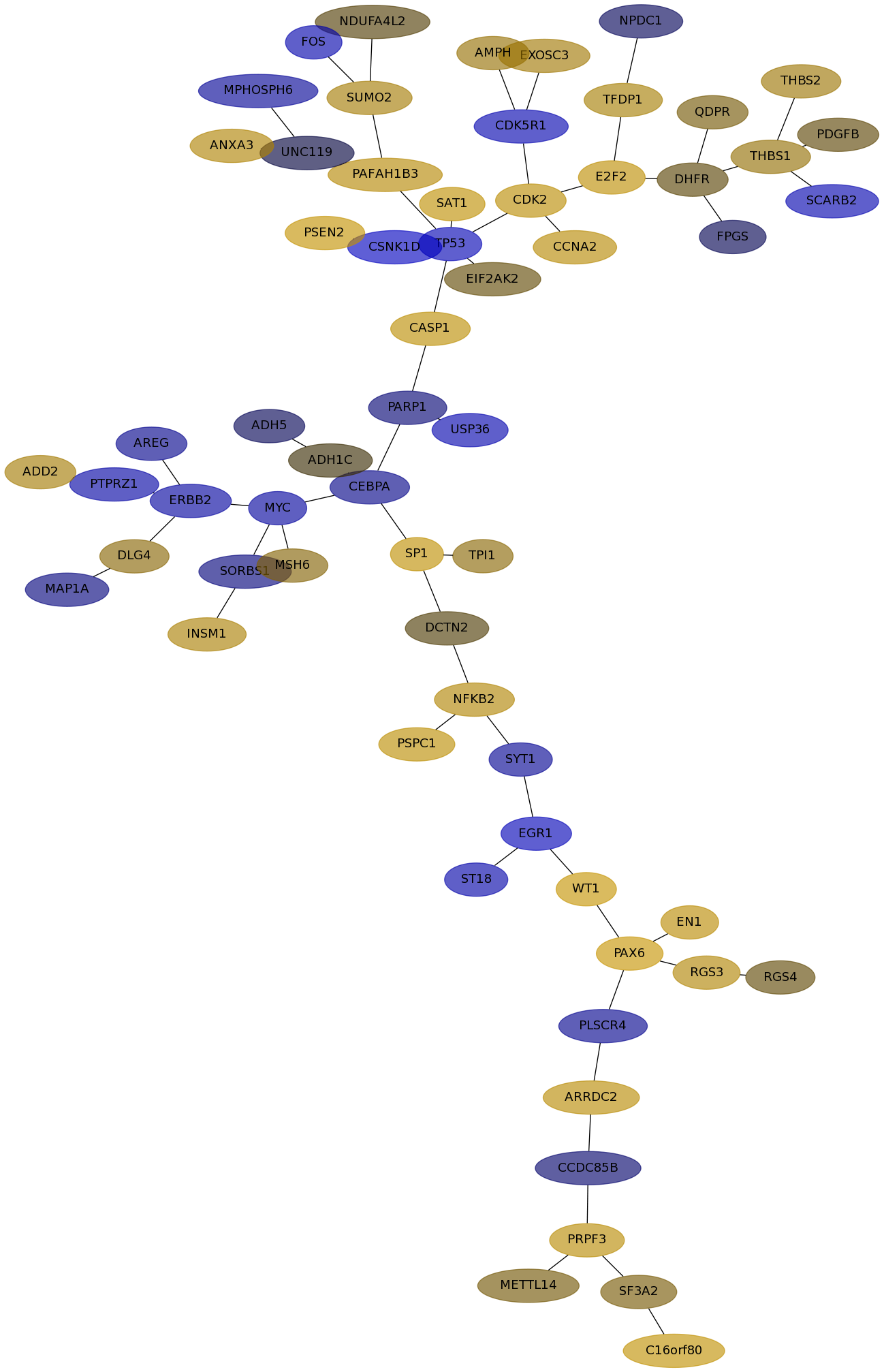
Score for each gene in subnetwork 6222 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| RGS4 |   | 4 | 440 | 842 | 831 | 0.022 | 0.160 | -0.006 | undef | 0.371 |
|---|
| ANXA3 |   | 3 | 557 | 141 | 170 | 0.152 | 0.132 | 0.194 | 0.090 | 0.030 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| EXOSC3 |   | 3 | 557 | 552 | 558 | 0.107 | 0.121 | undef | -0.074 | undef |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CEBPA |   | 3 | 557 | 1461 | 1440 | -0.058 | 0.005 | -0.100 | -0.199 | 0.039 |
|---|
| PDGFB |   | 7 | 256 | 1165 | 1135 | 0.020 | 0.046 | 0.085 | -0.049 | 0.338 |
|---|
| THBS1 |   | 5 | 360 | 1141 | 1109 | 0.078 | 0.044 | 0.185 | 0.200 | 0.267 |
|---|
| ADH5 |   | 1 | 1195 | 1928 | 1939 | -0.018 | -0.007 | 0.326 | 0.067 | 0.029 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| EIF2AK2 |   | 9 | 196 | 648 | 629 | 0.023 | -0.022 | -0.134 | 0.096 | -0.303 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| FPGS |   | 1 | 1195 | 1928 | 1939 | -0.016 | 0.016 | 0.175 | 0.068 | -0.130 |
|---|
| AMPH |   | 7 | 256 | 83 | 102 | 0.082 | 0.000 | -0.104 | 0.024 | 0.241 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| METTL14 |   | 2 | 743 | 1928 | 1911 | 0.035 | -0.039 | undef | undef | undef |
|---|
| RGS3 |   | 2 | 743 | 1846 | 1832 | 0.153 | 0.080 | 0.128 | undef | 0.037 |
|---|
| QDPR |   | 1 | 1195 | 1928 | 1939 | 0.037 | -0.085 | -0.158 | 0.092 | -0.045 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| ARRDC2 |   | 4 | 440 | 696 | 689 | 0.192 | -0.100 | undef | undef | undef |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| CASP1 |   | 6 | 301 | 552 | 544 | 0.204 | -0.166 | -0.046 | undef | -0.034 |
|---|
| MPHOSPH6 |   | 2 | 743 | 1846 | 1832 | -0.084 | 0.175 | -0.269 | 0.082 | 0.146 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| NDUFA4L2 |   | 2 | 743 | 1928 | 1911 | 0.015 | -0.030 | 0.065 | 0.139 | 0.174 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| THBS2 |   | 3 | 557 | 1903 | 1868 | 0.094 | 0.221 | 0.060 | 0.241 | 0.154 |
|---|
| PSPC1 |   | 6 | 301 | 991 | 964 | 0.203 | -0.289 | 0.298 | 0.148 | -0.107 |
|---|
| SAT1 |   | 6 | 301 | 236 | 239 | 0.219 | 0.010 | -0.066 | -0.119 | 0.063 |
|---|
| DLG4 |   | 5 | 360 | 806 | 784 | 0.059 | -0.113 | 0.151 | -0.131 | 0.123 |
|---|
| PRPF3 |   | 2 | 743 | 1928 | 1911 | 0.186 | -0.006 | 0.053 | 0.048 | -0.058 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| NPDC1 |   | 3 | 557 | 727 | 728 | -0.019 | 0.221 | 0.044 | 0.006 | -0.146 |
|---|
| SF3A2 |   | 3 | 557 | 1203 | 1187 | 0.038 | -0.181 | 0.094 | -0.172 | -0.067 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| USP36 |   | 3 | 557 | 882 | 871 | -0.144 | -0.044 | 0.203 | undef | 0.014 |
|---|
| ADH1C |   | 2 | 743 | 1629 | 1614 | 0.009 | 0.062 | 0.163 | 0.059 | -0.024 |
|---|
| SCARB2 |   | 1 | 1195 | 1928 | 1939 | -0.152 | 0.027 | 0.030 | 0.237 | -0.073 |
|---|
| PAFAH1B3 |   | 2 | 743 | 1928 | 1911 | 0.172 | -0.018 | -0.104 | 0.060 | -0.024 |
|---|
| AREG |   | 5 | 360 | 1141 | 1109 | -0.067 | 0.112 | 0.076 | -0.173 | -0.054 |
|---|
| TFDP1 |   | 6 | 301 | 806 | 783 | 0.123 | -0.178 | 0.129 | undef | -0.065 |
|---|
| CCDC85B |   | 3 | 557 | 806 | 792 | -0.030 | -0.293 | 0.123 | undef | 0.115 |
|---|
| DHFR |   | 4 | 440 | 179 | 209 | 0.019 | 0.029 | 0.232 | 0.129 | -0.050 |
|---|
| C16orf80 |   | 1 | 1195 | 1928 | 1939 | 0.227 | -0.070 | -0.030 | 0.023 | 0.076 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| TPI1 |   | 1 | 1195 | 1928 | 1939 | 0.061 | -0.121 | -0.057 | undef | 0.069 |
|---|
| CSNK1D |   | 6 | 301 | 614 | 602 | -0.222 | -0.044 | 0.170 | -0.088 | 0.013 |
|---|
GO Enrichment output for subnetwork 6222 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycine metabolic process | GO:0006544 |  | 1.246E-10 | 3.043E-07 |
|---|
| one-carbon metabolic process | GO:0006730 |  | 2.28E-10 | 2.785E-07 |
|---|
| glycine biosynthetic process | GO:0006545 |  | 1.873E-09 | 1.525E-06 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 8.509E-09 | 5.197E-06 |
|---|
| regulation of helicase activity | GO:0051095 |  | 1.302E-08 | 6.361E-06 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.318E-08 | 5.368E-06 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 3.634E-07 | 1.268E-04 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 8.562E-07 | 2.615E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 8.562E-07 | 2.324E-04 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 2.977E-06 | 7.273E-04 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 4.748E-06 | 1.054E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycine metabolic process | GO:0006544 |  | 1.841E-10 | 4.428E-07 |
|---|
| one-carbon metabolic process | GO:0006730 |  | 5.268E-10 | 6.337E-07 |
|---|
| glycine biosynthetic process | GO:0006545 |  | 3.189E-09 | 2.558E-06 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 1.033E-08 | 6.211E-06 |
|---|
| regulation of helicase activity | GO:0051095 |  | 2.215E-08 | 1.066E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.639E-08 | 1.459E-05 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 4.418E-07 | 1.519E-04 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 1.276E-06 | 3.838E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.276E-06 | 3.411E-04 |
|---|
| negative regulation of DNA recombination | GO:0045910 |  | 4.433E-06 | 1.067E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 7.066E-06 | 1.546E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycine metabolic process | GO:0006544 |  | 1.938E-10 | 4.457E-07 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 1.783E-08 | 2.05E-05 |
|---|
| one-carbon metabolic process | GO:0006730 |  | 3.692E-08 | 2.831E-05 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 5.464E-07 | 3.142E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.351E-06 | 6.217E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.675E-06 | 1.025E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 5.325E-06 | 1.75E-03 |
|---|
| synaptic vesicle endocytosis | GO:0048488 |  | 9.274E-06 | 2.666E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.477E-05 | 3.774E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.478E-05 | 3.4E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 3.135E-05 | 6.554E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 7.105E-07 | 1.31E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.426E-06 | 1.314E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.592E-06 | 1.592E-03 |
|---|
| one-carbon metabolic process | GO:0006730 |  | 6.769E-06 | 3.119E-03 |
|---|
| glycine metabolic process | GO:0006544 |  | 1.431E-05 | 5.276E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.431E-05 | 4.397E-03 |
|---|
| regulation of blood vessel endothelial cell migration | GO:0043535 |  | 2.137E-05 | 5.627E-03 |
|---|
| learning or memory | GO:0007611 |  | 2.512E-05 | 5.786E-03 |
|---|
| synaptic vesicle transport | GO:0048489 |  | 3.039E-05 | 6.224E-03 |
|---|
| positive regulation of DNA replication | GO:0045740 |  | 7.145E-05 | 0.01316824 |
|---|
| regulation of endothelial cell migration | GO:0010594 |  | 7.145E-05 | 0.01197113 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycine metabolic process | GO:0006544 |  | 1.938E-10 | 4.457E-07 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 1.783E-08 | 2.05E-05 |
|---|
| one-carbon metabolic process | GO:0006730 |  | 3.692E-08 | 2.831E-05 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 5.464E-07 | 3.142E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.351E-06 | 6.217E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.675E-06 | 1.025E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 5.325E-06 | 1.75E-03 |
|---|
| synaptic vesicle endocytosis | GO:0048488 |  | 9.274E-06 | 2.666E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.477E-05 | 3.774E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.478E-05 | 3.4E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 3.135E-05 | 6.554E-03 |
|---|



