Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6220
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7598 | 3.156e-03 | 4.980e-04 | 7.545e-01 | 1.186e-06 |
|---|
| Loi | 0.2282 | 8.364e-02 | 1.227e-02 | 4.363e-01 | 4.476e-04 |
|---|
| Schmidt | 0.6718 | 0.000e+00 | 0.000e+00 | 4.662e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.497e-03 | 0.000e+00 |
|---|
| Wang | 0.2592 | 3.012e-03 | 4.678e-02 | 4.098e-01 | 5.775e-05 |
|---|
Expression data for subnetwork 6220 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
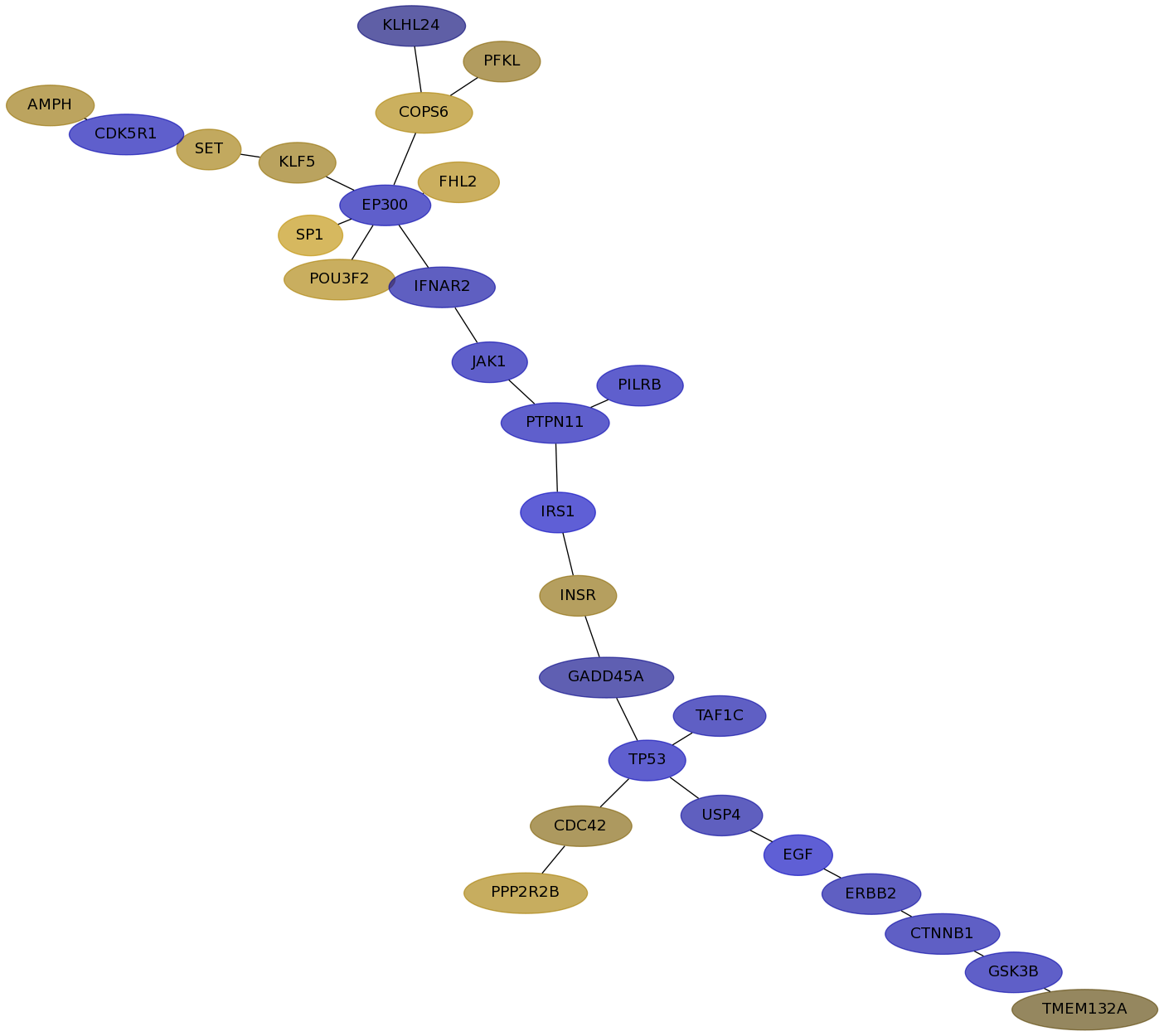
Score for each gene in subnetwork 6220 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| KLHL24 |   | 4 | 440 | 682 | 678 | -0.036 | -0.011 | 0.062 | 0.044 | -0.125 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| IFNAR2 |   | 4 | 440 | 1558 | 1528 | -0.100 | 0.055 | 0.106 | -0.294 | -0.058 |
|---|
| USP4 |   | 7 | 256 | 1 | 47 | -0.093 | 0.147 | 0.097 | -0.392 | 0.054 |
|---|
| PILRB |   | 1 | 1195 | 1999 | 2004 | -0.154 | 0.243 | -0.005 | undef | -0.146 |
|---|
| TAF1C |   | 1 | 1195 | 1999 | 2004 | -0.111 | 0.132 | -0.069 | -0.041 | 0.034 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| TMEM132A |   | 1 | 1195 | 1999 | 2004 | 0.019 | 0.127 | -0.141 | undef | -0.092 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| JAK1 |   | 5 | 360 | 696 | 680 | -0.140 | 0.034 | -0.176 | 0.111 | 0.159 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| AMPH |   | 7 | 256 | 83 | 102 | 0.082 | 0.000 | -0.104 | 0.024 | 0.241 |
|---|
| SET |   | 1 | 1195 | 1999 | 2004 | 0.103 | 0.250 | 0.196 | undef | -0.128 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
GO Enrichment output for subnetwork 6220 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.643E-13 | 1.867E-09 |
|---|
| organ growth | GO:0035265 |  | 6.367E-10 | 7.777E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 6.367E-10 | 5.185E-07 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.145E-09 | 6.996E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.271E-09 | 6.212E-07 |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.271E-09 | 5.177E-07 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.271E-09 | 4.437E-07 |
|---|
| regulation of histone modification | GO:0031056 |  | 2.285E-09 | 6.977E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 2.285E-09 | 6.202E-07 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.843E-09 | 6.946E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.965E-09 | 1.325E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.139E-12 | 2.741E-09 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 8.783E-10 | 1.057E-06 |
|---|
| organ growth | GO:0035265 |  | 8.783E-10 | 7.044E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 8.783E-10 | 5.283E-07 |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.754E-09 | 8.438E-07 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.754E-09 | 7.032E-07 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.964E-09 | 6.751E-07 |
|---|
| regulation of histone modification | GO:0031056 |  | 3.151E-09 | 9.476E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.151E-09 | 8.423E-07 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 4.227E-09 | 1.017E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.242E-09 | 1.147E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.144E-12 | 2.631E-09 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.472E-10 | 3.992E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.42E-09 | 1.855E-06 |
|---|
| organ growth | GO:0035265 |  | 2.42E-09 | 1.391E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.42E-09 | 1.113E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 4.828E-09 | 1.851E-06 |
|---|
| nucleosome disassembly | GO:0006337 |  | 4.828E-09 | 1.586E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 5.599E-09 | 1.61E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 8.672E-09 | 2.216E-06 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 9.708E-09 | 2.233E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 2.054E-08 | 4.295E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.715E-07 | 3.161E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.874E-07 | 4.492E-04 |
|---|
| ER overload response | GO:0006983 |  | 4.874E-07 | 2.994E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.874E-07 | 2.246E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 7.15E-07 | 2.636E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.875E-06 | 2.112E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 6.875E-06 | 1.81E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 9.635E-06 | 2.22E-03 |
|---|
| response to hormone stimulus | GO:0009725 |  | 9.926E-06 | 2.033E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.089E-05 | 2.008E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.089E-05 | 1.825E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.144E-12 | 2.631E-09 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.472E-10 | 3.992E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.42E-09 | 1.855E-06 |
|---|
| organ growth | GO:0035265 |  | 2.42E-09 | 1.391E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.42E-09 | 1.113E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 4.828E-09 | 1.851E-06 |
|---|
| nucleosome disassembly | GO:0006337 |  | 4.828E-09 | 1.586E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 5.599E-09 | 1.61E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 8.672E-09 | 2.216E-06 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 9.708E-09 | 2.233E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 2.054E-08 | 4.295E-06 |
|---|



