Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6130
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7612 | 3.079e-03 | 4.820e-04 | 7.506e-01 | 1.114e-06 |
|---|
| Loi | 0.2257 | 8.755e-02 | 1.336e-02 | 4.456e-01 | 5.211e-04 |
|---|
| Schmidt | 0.6731 | 0.000e+00 | 0.000e+00 | 4.523e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7902 | 0.000e+00 | 0.000e+00 | 3.547e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.031e-03 | 4.695e-02 | 4.106e-01 | 5.844e-05 |
|---|
Expression data for subnetwork 6130 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
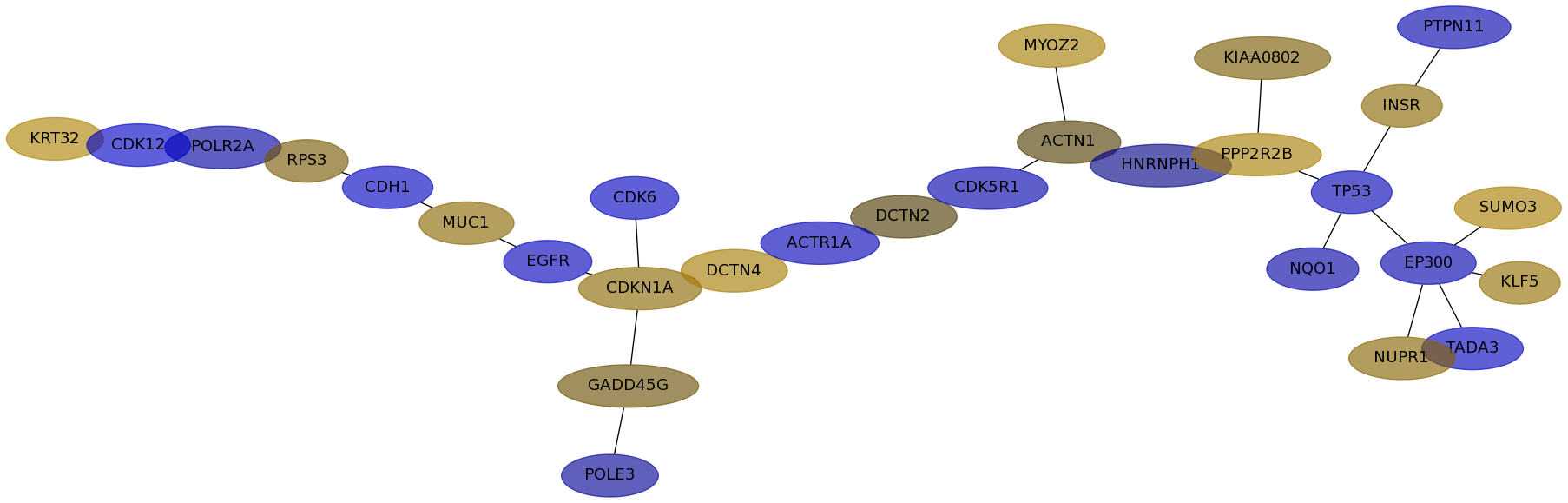
Score for each gene in subnetwork 6130 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| MYOZ2 |   | 1 | 1195 | 2255 | 2257 | 0.122 | 0.041 | -0.081 | -0.115 | 0.059 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| SUMO3 |   | 3 | 557 | 412 | 430 | 0.126 | 0.081 | 0.087 | 0.138 | -0.073 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| ACTR1A |   | 2 | 743 | 2255 | 2227 | -0.184 | 0.178 | 0.121 | 0.197 | 0.100 |
|---|
| POLR2A |   | 6 | 301 | 1547 | 1510 | -0.107 | 0.247 | 0.091 | 0.040 | 0.141 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| TADA3 |   | 1 | 1195 | 2255 | 2257 | -0.217 | 0.083 | -0.076 | 0.097 | -0.109 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| NUPR1 |   | 3 | 557 | 1222 | 1210 | 0.057 | -0.031 | -0.241 | 0.145 | 0.013 |
|---|
| KIAA0802 |   | 3 | 557 | 2046 | 2009 | 0.041 | -0.123 | 0.123 | 0.059 | 0.037 |
|---|
| DCTN4 |   | 5 | 360 | 1010 | 987 | 0.122 | -0.025 | -0.111 | 0.173 | -0.105 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| RPS3 |   | 2 | 743 | 1903 | 1878 | 0.038 | 0.007 | 0.236 | -0.194 | 0.006 |
|---|
| KRT32 |   | 1 | 1195 | 2255 | 2257 | 0.143 | 0.072 | 0.162 | -0.201 | -0.158 |
|---|
| CDK12 |   | 1 | 1195 | 2255 | 2257 | -0.258 | 0.130 | -0.073 | 0.295 | -0.006 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
GO Enrichment output for subnetwork 6130 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.471E-10 | 1.825E-06 |
|---|
| organ growth | GO:0035265 |  | 7.471E-10 | 9.126E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 7.471E-10 | 6.084E-07 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.492E-09 | 9.11E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 2.681E-09 | 1.31E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.377E-08 | 5.608E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.119E-08 | 7.395E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 2.426E-08 | 7.409E-06 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.566E-08 | 9.681E-06 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 4.996E-08 | 1.221E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 5.708E-08 | 1.268E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.168E-09 | 2.811E-06 |
|---|
| organ growth | GO:0035265 |  | 1.168E-09 | 1.406E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.168E-09 | 9.371E-07 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.333E-09 | 1.403E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 4.191E-09 | 2.017E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.756E-08 | 7.04E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.773E-08 | 9.531E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 3.678E-08 | 1.106E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.186E-08 | 1.654E-05 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 7.799E-08 | 1.877E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 9.892E-08 | 2.164E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 4.055E-10 | 9.327E-07 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.214E-09 | 1.396E-06 |
|---|
| organ growth | GO:0035265 |  | 2.826E-09 | 2.166E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.826E-09 | 1.625E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.638E-09 | 2.594E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.718E-08 | 1.425E-05 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 5.169E-08 | 1.698E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 6.046E-08 | 1.738E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.038E-08 | 1.799E-05 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.082E-07 | 2.489E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.814E-07 | 3.793E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.843E-06 | 3.397E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 5.293E-06 | 4.877E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.226E-06 | 4.439E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 7.651E-06 | 3.525E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 8.569E-06 | 3.159E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 8.858E-06 | 2.721E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 9.134E-06 | 2.405E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 9.134E-06 | 2.104E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 9.769E-06 | 2E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.412E-05 | 2.602E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.821E-05 | 3.05E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 4.055E-10 | 9.327E-07 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.214E-09 | 1.396E-06 |
|---|
| organ growth | GO:0035265 |  | 2.826E-09 | 2.166E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.826E-09 | 1.625E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.638E-09 | 2.594E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.718E-08 | 1.425E-05 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 5.169E-08 | 1.698E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 6.046E-08 | 1.738E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.038E-08 | 1.799E-05 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.082E-07 | 2.489E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.814E-07 | 3.793E-05 |
|---|



