Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6129
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7613 | 3.076e-03 | 4.820e-04 | 7.504e-01 | 1.113e-06 |
|---|
| Loi | 0.2257 | 8.762e-02 | 1.338e-02 | 4.457e-01 | 5.226e-04 |
|---|
| Schmidt | 0.6732 | 0.000e+00 | 0.000e+00 | 4.516e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7902 | 0.000e+00 | 0.000e+00 | 3.548e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.030e-03 | 4.694e-02 | 4.106e-01 | 5.840e-05 |
|---|
Expression data for subnetwork 6129 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
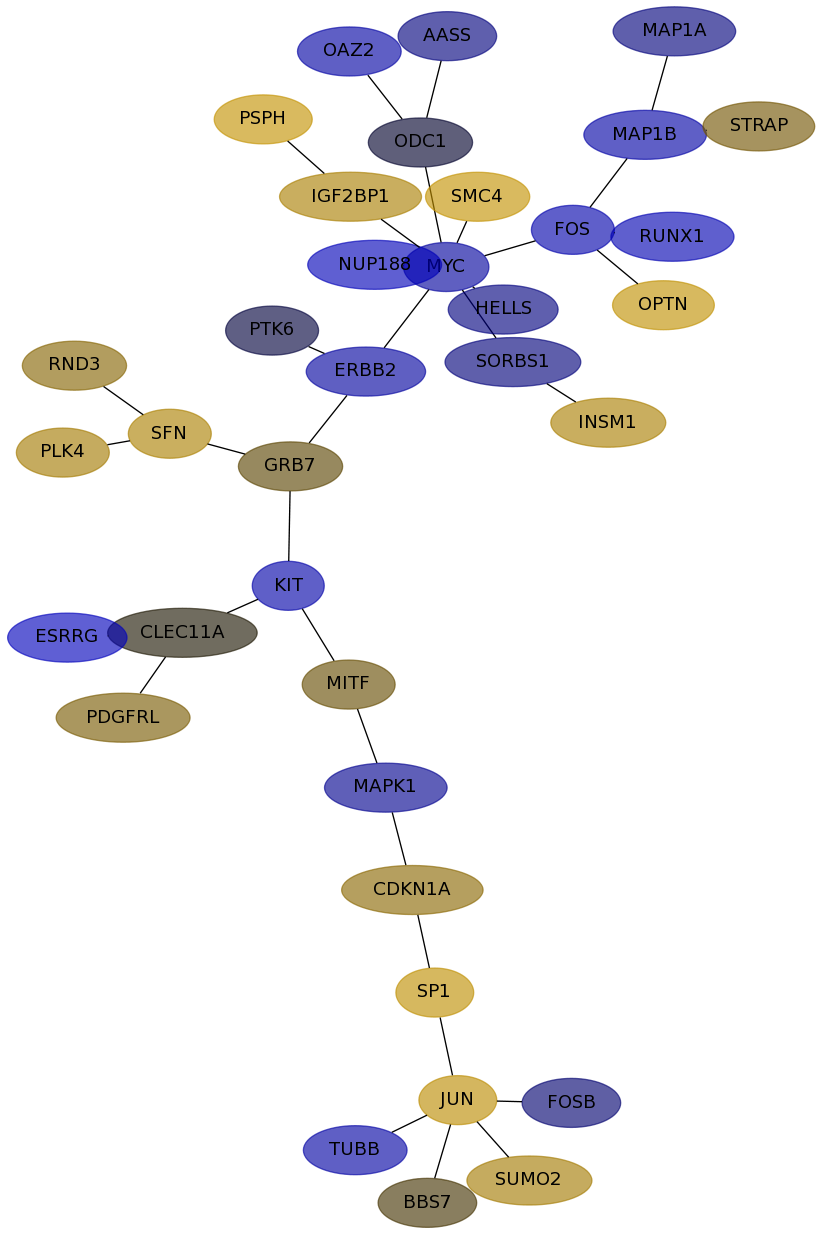
Score for each gene in subnetwork 6129 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PTK6 |   | 3 | 557 | 2141 | 2102 | -0.010 | 0.204 | -0.343 | 0.069 | 0.213 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NUP188 |   | 8 | 222 | 1116 | 1079 | -0.187 | 0.094 | 0.058 | 0.161 | -0.070 |
|---|
| AASS |   | 2 | 743 | 2147 | 2126 | -0.047 | 0.229 | 0.021 | 0.187 | 0.002 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PDGFRL |   | 1 | 1195 | 2174 | 2178 | 0.042 | 0.110 | -0.038 | 0.119 | 0.143 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BBS7 |   | 1 | 1195 | 2174 | 2178 | 0.012 | -0.065 | 0.263 | -0.059 | 0.101 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| FOSB |   | 5 | 360 | 1534 | 1501 | -0.033 | 0.046 | 0.129 | -0.104 | 0.095 |
|---|
| MITF |   | 5 | 360 | 696 | 680 | 0.027 | 0.032 | 0.065 | 0.145 | 0.183 |
|---|
| PLK4 |   | 2 | 743 | 957 | 958 | 0.114 | -0.154 | 0.260 | 0.136 | 0.027 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| STRAP |   | 7 | 256 | 412 | 414 | 0.035 | -0.128 | 0.074 | 0.154 | -0.049 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| SMC4 |   | 3 | 557 | 83 | 117 | 0.247 | -0.111 | 0.214 | 0.072 | 0.094 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| ESRRG |   | 6 | 301 | 614 | 602 | -0.205 | 0.197 | -0.007 | 0.155 | -0.085 |
|---|
| ODC1 |   | 4 | 440 | 141 | 165 | -0.006 | -0.099 | 0.209 | -0.207 | -0.107 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| HELLS |   | 2 | 743 | 1846 | 1832 | -0.050 | -0.018 | 0.199 | 0.057 | 0.107 |
|---|
| RND3 |   | 2 | 743 | 1346 | 1333 | 0.056 | 0.154 | 0.167 | -0.018 | 0.218 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| OAZ2 |   | 1 | 1195 | 2174 | 2178 | -0.112 | -0.023 | 0.215 | -0.061 | 0.252 |
|---|
| RUNX1 |   | 5 | 360 | 906 | 890 | -0.161 | 0.140 | 0.016 | 0.102 | 0.025 |
|---|
| MAPK1 |   | 1 | 1195 | 2174 | 2178 | -0.072 | 0.006 | 0.118 | -0.034 | 0.043 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| TUBB |   | 5 | 360 | 614 | 612 | -0.118 | 0.170 | 0.200 | undef | -0.045 |
|---|
GO Enrichment output for subnetwork 6129 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.191E-10 | 5.353E-07 |
|---|
| pigmentation | GO:0043473 |  | 2.345E-09 | 2.864E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 9.111E-09 | 7.419E-06 |
|---|
| spindle organization | GO:0007051 |  | 2.094E-08 | 1.279E-05 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.139E-08 | 1.045E-05 |
|---|
| cell killing | GO:0001906 |  | 5.863E-08 | 2.387E-05 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 3.448E-07 | 1.203E-04 |
|---|
| positive regulation of myeloid leukocyte differentiation | GO:0002763 |  | 1.44E-06 | 4.398E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 2.053E-06 | 5.574E-04 |
|---|
| L-serine metabolic process | GO:0006563 |  | 2.818E-06 | 6.884E-04 |
|---|
| polyamine metabolic process | GO:0006595 |  | 3.75E-06 | 8.329E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 3.24E-10 | 7.796E-07 |
|---|
| pigmentation | GO:0043473 |  | 4.164E-09 | 5.009E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.346E-08 | 1.079E-05 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 3.159E-08 | 1.9E-05 |
|---|
| spindle organization | GO:0007051 |  | 3.39E-08 | 1.631E-05 |
|---|
| cell killing | GO:0001906 |  | 8.653E-08 | 3.47E-05 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 2.319E-07 | 7.969E-05 |
|---|
| L-serine metabolic process | GO:0006563 |  | 2.753E-06 | 8.281E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 2.753E-06 | 7.361E-04 |
|---|
| polyamine metabolic process | GO:0006595 |  | 5.027E-06 | 1.209E-03 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 6.521E-06 | 1.426E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 8.677E-10 | 1.996E-06 |
|---|
| pigmentation | GO:0043473 |  | 5.247E-09 | 6.034E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.162E-08 | 1.658E-05 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.632E-08 | 3.238E-05 |
|---|
| spindle organization | GO:0007051 |  | 8.057E-08 | 3.706E-05 |
|---|
| cell killing | GO:0001906 |  | 1.694E-07 | 6.494E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 6.762E-07 | 2.222E-04 |
|---|
| polyamine metabolic process | GO:0006595 |  | 5.779E-06 | 1.661E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 5.779E-06 | 1.477E-03 |
|---|
| cellular pigmentation | GO:0033059 |  | 1.366E-05 | 3.142E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.438E-05 | 3.007E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyamine metabolic process | GO:0006595 |  | 2.65E-06 | 4.884E-03 |
|---|
| pigmentation during development | GO:0048066 |  | 3.965E-06 | 3.654E-03 |
|---|
| pigmentation | GO:0043473 |  | 3.146E-05 | 0.01932843 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 6.092E-05 | 0.02806992 |
|---|
| sister chromatid segregation | GO:0000819 |  | 7.036E-05 | 0.02593645 |
|---|
| endocrine pancreas development | GO:0031018 |  | 1.364E-04 | 0.04189285 |
|---|
| melanocyte differentiation | GO:0030318 |  | 1.364E-04 | 0.03590815 |
|---|
| DNA methylation | GO:0006306 |  | 1.364E-04 | 0.03141964 |
|---|
| pyrimidine base metabolic process | GO:0006206 |  | 1.364E-04 | 0.02792856 |
|---|
| B cell receptor signaling pathway | GO:0050853 |  | 1.364E-04 | 0.02513571 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.041E-04 | 0.03419627 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 8.677E-10 | 1.996E-06 |
|---|
| pigmentation | GO:0043473 |  | 5.247E-09 | 6.034E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.162E-08 | 1.658E-05 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.632E-08 | 3.238E-05 |
|---|
| spindle organization | GO:0007051 |  | 8.057E-08 | 3.706E-05 |
|---|
| cell killing | GO:0001906 |  | 1.694E-07 | 6.494E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 6.762E-07 | 2.222E-04 |
|---|
| polyamine metabolic process | GO:0006595 |  | 5.779E-06 | 1.661E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 5.779E-06 | 1.477E-03 |
|---|
| cellular pigmentation | GO:0033059 |  | 1.366E-05 | 3.142E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.438E-05 | 3.007E-03 |
|---|



