Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6121
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7614 | 3.071e-03 | 4.810e-04 | 7.501e-01 | 1.108e-06 |
|---|
| Loi | 0.2255 | 8.791e-02 | 1.346e-02 | 4.464e-01 | 5.284e-04 |
|---|
| Schmidt | 0.6732 | 0.000e+00 | 0.000e+00 | 4.519e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7901 | 0.000e+00 | 0.000e+00 | 3.553e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.025e-03 | 4.689e-02 | 4.103e-01 | 5.821e-05 |
|---|
Expression data for subnetwork 6121 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
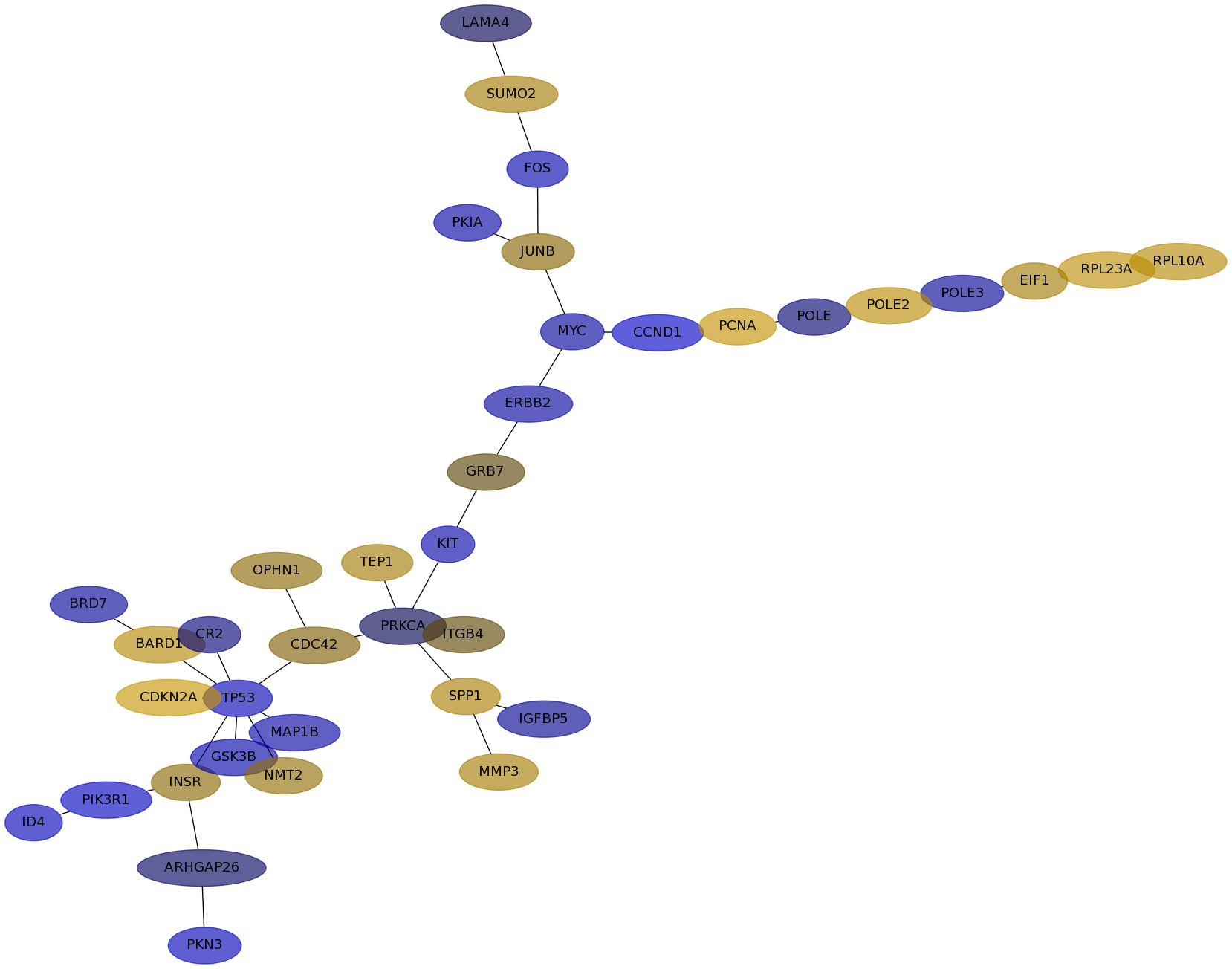
Score for each gene in subnetwork 6121 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| OPHN1 |   | 2 | 743 | 1822 | 1806 | 0.062 | 0.278 | 0.194 | 0.008 | -0.182 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RPL23A |   | 5 | 360 | 842 | 828 | 0.208 | -0.007 | 0.024 | -0.242 | 0.055 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| EIF1 |   | 1 | 1195 | 2282 | 2283 | 0.114 | -0.046 | 0.187 | -0.002 | 0.137 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| TEP1 |   | 2 | 743 | 1967 | 1945 | 0.111 | 0.126 | -0.039 | 0.182 | 0.134 |
|---|
| CR2 |   | 5 | 360 | 957 | 938 | -0.042 | -0.089 | 0.101 | 0.018 | 0.015 |
|---|
| RPL10A |   | 2 | 743 | 1785 | 1775 | 0.176 | 0.061 | 0.219 | -0.063 | -0.024 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NMT2 |   | 7 | 256 | 906 | 876 | 0.077 | -0.296 | 0.134 | -0.124 | 0.217 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| MMP3 |   | 6 | 301 | 179 | 205 | 0.121 | 0.171 | 0.081 | 0.198 | 0.192 |
|---|
| POLE |   | 1 | 1195 | 2282 | 2283 | -0.036 | -0.107 | 0.144 | -0.026 | 0.009 |
|---|
| POLE2 |   | 4 | 440 | 727 | 723 | 0.200 | -0.203 | 0.049 | 0.000 | -0.010 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
GO Enrichment output for subnetwork 6121 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of embryonic development | GO:0045995 |  | 1.917E-08 | 4.683E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.046E-07 | 1.277E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.78E-07 | 1.45E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.993E-07 | 1.217E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.76E-07 | 1.349E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.053E-07 | 1.243E-04 |
|---|
| nuclear migration | GO:0007097 |  | 4.295E-07 | 1.499E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.295E-07 | 1.312E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.94E-07 | 1.612E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 5.94E-07 | 1.451E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.001E-07 | 1.333E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of embryonic development | GO:0045995 |  | 2.765E-08 | 6.653E-05 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.507E-07 | 1.813E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.791E-07 | 2.238E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.123E-07 | 1.879E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.883E-07 | 1.868E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.974E-07 | 1.594E-04 |
|---|
| nuclear migration | GO:0007097 |  | 5.663E-07 | 1.946E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.663E-07 | 1.703E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.755E-07 | 2.073E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 8.494E-07 | 2.044E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.634E-07 | 1.889E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.636E-07 | 1.066E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 6.961E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 5.105E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 3.829E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.746E-07 | 3.563E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.77E-07 | 3.362E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.328E-06 | 4.363E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.328E-06 | 3.818E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 3.393E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.501E-06 | 3.452E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.207E-06 | 4.615E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ER overload response | GO:0006983 |  | 8.418E-07 | 1.551E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 8.418E-07 | 7.757E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.63E-06 | 1.001E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.819E-06 | 8.379E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.25E-06 | 8.293E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.995E-06 | 1.227E-03 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.995E-06 | 1.052E-03 |
|---|
| negative regulation of nucleocytoplasmic transport | GO:0046823 |  | 5.002E-06 | 1.152E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.024E-05 | 2.097E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.184E-05 | 2.182E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.184E-05 | 1.983E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 4.636E-07 | 1.066E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 6.961E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 5.105E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 3.829E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.746E-07 | 3.563E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.77E-07 | 3.362E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.328E-06 | 4.363E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.328E-06 | 3.818E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 3.393E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.501E-06 | 3.452E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.207E-06 | 4.615E-04 |
|---|



