Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6110
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7616 | 3.056e-03 | 4.780e-04 | 7.494e-01 | 1.095e-06 |
|---|
| Loi | 0.2251 | 8.865e-02 | 1.367e-02 | 4.481e-01 | 5.433e-04 |
|---|
| Schmidt | 0.6734 | 0.000e+00 | 0.000e+00 | 4.492e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7901 | 0.000e+00 | 0.000e+00 | 3.552e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.033e-03 | 4.696e-02 | 4.107e-01 | 5.849e-05 |
|---|
Expression data for subnetwork 6110 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
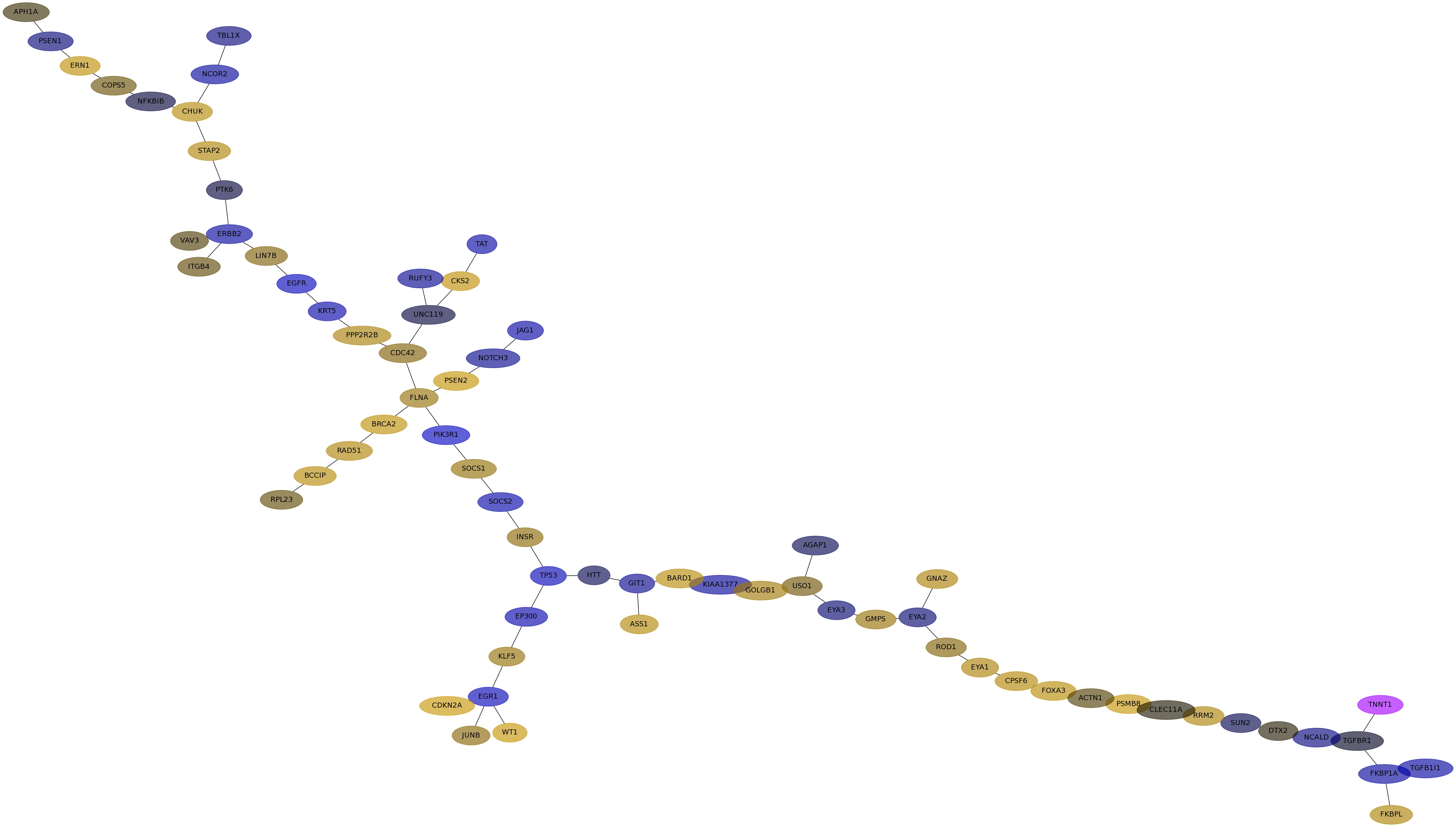
Score for each gene in subnetwork 6110 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PTK6 |   | 3 | 557 | 2141 | 2102 | -0.010 | 0.204 | -0.343 | 0.069 | 0.213 |
|---|
| BCCIP |   | 1 | 1195 | 2270 | 2271 | 0.176 | -0.138 | 0.119 | undef | -0.156 |
|---|
| SOCS2 |   | 3 | 557 | 1688 | 1660 | -0.130 | -0.028 | 0.051 | -0.171 | 0.027 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ASS1 |   | 1 | 1195 | 2270 | 2271 | 0.160 | -0.094 | -0.015 | undef | -0.052 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHUK |   | 6 | 301 | 318 | 330 | 0.173 | 0.080 | 0.152 | undef | 0.215 |
|---|
| GIT1 |   | 2 | 743 | 1510 | 1502 | -0.067 | -0.099 | -0.045 | undef | -0.065 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| JAG1 |   | 1 | 1195 | 2270 | 2271 | -0.116 | 0.319 | 0.138 | 0.029 | 0.203 |
|---|
| PSMB8 |   | 3 | 557 | 928 | 925 | 0.254 | -0.071 | -0.088 | undef | -0.139 |
|---|
| LIN7B |   | 1 | 1195 | 2270 | 2271 | 0.047 | -0.123 | -0.141 | undef | -0.147 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| SOCS1 |   | 3 | 557 | 1717 | 1689 | 0.074 | -0.153 | -0.177 | -0.202 | -0.100 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| TNNT1 |   | 3 | 557 | 1914 | 1877 | undef | undef | undef | 0.145 | undef |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| NOTCH3 |   | 5 | 360 | 806 | 784 | -0.063 | 0.115 | 0.136 | undef | -0.082 |
|---|
| CPSF6 |   | 6 | 301 | 318 | 330 | 0.164 | 0.052 | 0.091 | 0.129 | -0.088 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| USO1 |   | 2 | 743 | 1688 | 1673 | 0.031 | -0.003 | -0.027 | 0.096 | -0.077 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| APH1A |   | 3 | 557 | 1803 | 1777 | 0.009 | -0.165 | 0.107 | 0.183 | -0.098 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| RRM2 |   | 1 | 1195 | 2270 | 2271 | 0.140 | -0.135 | -0.094 | undef | 0.130 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| COPS5 |   | 2 | 743 | 1131 | 1136 | 0.028 | -0.117 | -0.030 | 0.155 | 0.108 |
|---|
| NCALD |   | 4 | 440 | 1709 | 1672 | -0.046 | 0.078 | 0.090 | undef | 0.004 |
|---|
| ERN1 |   | 1 | 1195 | 2270 | 2271 | 0.205 | -0.027 | 0.000 | 0.029 | -0.092 |
|---|
| ROD1 |   | 4 | 440 | 1564 | 1542 | 0.049 | 0.088 | 0.052 | 0.011 | -0.223 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| STAP2 |   | 1 | 1195 | 2270 | 2271 | 0.148 | -0.010 | -0.165 | -0.152 | -0.237 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| EYA1 |   | 8 | 222 | 1010 | 981 | 0.135 | -0.065 | 0.162 | 0.068 | -0.088 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| TGFBR1 |   | 4 | 440 | 1793 | 1755 | -0.004 | 0.036 | -0.078 | 0.181 | undef |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| FKBP1A |   | 1 | 1195 | 2270 | 2271 | -0.092 | 0.077 | -0.095 | 0.088 | -0.110 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| RPL23 |   | 2 | 743 | 2215 | 2193 | 0.024 | -0.022 | 0.164 | 0.086 | -0.032 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| GMPS |   | 7 | 256 | 318 | 328 | 0.083 | -0.022 | 0.044 | -0.019 | 0.151 |
|---|
| SUN2 |   | 1 | 1195 | 2270 | 2271 | -0.015 | -0.031 | undef | undef | undef |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| AGAP1 |   | 1 | 1195 | 2270 | 2271 | -0.015 | -0.061 | -0.070 | 0.157 | -0.124 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| FKBPL |   | 1 | 1195 | 2270 | 2271 | 0.139 | 0.094 | 0.017 | undef | 0.151 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| DTX2 |   | 2 | 743 | 1793 | 1779 | 0.005 | 0.174 | -0.122 | undef | -0.200 |
|---|
| TBL1X |   | 4 | 440 | 1735 | 1699 | -0.044 | 0.017 | -0.192 | 0.198 | -0.009 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| RUFY3 |   | 1 | 1195 | 2270 | 2271 | -0.079 | 0.281 | 0.116 | 0.163 | -0.088 |
|---|
| NFKBIB |   | 2 | 743 | 1938 | 1922 | -0.008 | -0.192 | -0.185 | -0.098 | -0.239 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| GOLGB1 |   | 3 | 557 | 983 | 979 | 0.104 | 0.352 | 0.044 | 0.079 | 0.175 |
|---|
GO Enrichment output for subnetwork 6110 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Notch receptor processing | GO:0007220 |  | 3.694E-09 | 9.025E-06 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 2.565E-08 | 3.133E-05 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 9.157E-08 | 7.457E-05 |
|---|
| Notch signaling pathway | GO:0007219 |  | 3.258E-07 | 1.99E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.553E-07 | 1.736E-04 |
|---|
| membrane protein proteolysis | GO:0033619 |  | 5.111E-07 | 2.081E-04 |
|---|
| glycoprotein catabolic process | GO:0006516 |  | 9.678E-07 | 3.378E-04 |
|---|
| nuclear migration | GO:0007097 |  | 2.833E-06 | 8.651E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.833E-06 | 7.69E-04 |
|---|
| regulation of cell projection organization | GO:0031344 |  | 7.375E-06 | 1.802E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.871E-06 | 1.748E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Notch receptor processing | GO:0007220 |  | 4.878E-09 | 1.174E-05 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 3.385E-08 | 4.072E-05 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 1.208E-07 | 9.686E-05 |
|---|
| Notch signaling pathway | GO:0007219 |  | 2.985E-07 | 1.796E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.682E-07 | 2.253E-04 |
|---|
| membrane protein proteolysis | GO:0033619 |  | 4.682E-07 | 1.878E-04 |
|---|
| glycoprotein catabolic process | GO:0006516 |  | 1.274E-06 | 4.38E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.49E-06 | 1.05E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.49E-06 | 9.331E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.433E-06 | 1.307E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.083E-06 | 1.33E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.224E-07 | 1.202E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 4.16E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 2.773E-03 |
|---|
| cell fate commitment | GO:0045165 |  | 4.505E-06 | 2.59E-03 |
|---|
| Notch signaling pathway | GO:0007219 |  | 4.902E-06 | 2.255E-03 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 7.197E-06 | 2.759E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.197E-06 | 2.365E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.197E-06 | 2.069E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.255E-06 | 1.854E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.253E-05 | 2.881E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.253E-05 | 2.619E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 8.705E-09 | 1.604E-05 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 6.034E-08 | 5.56E-05 |
|---|
| membrane protein ectodomain proteolysis | GO:0006509 |  | 6.034E-08 | 3.707E-05 |
|---|
| glycoprotein catabolic process | GO:0006516 |  | 8.327E-07 | 3.837E-04 |
|---|
| Notch signaling pathway | GO:0007219 |  | 9.635E-06 | 3.551E-03 |
|---|
| protein maturation | GO:0051604 |  | 1.996E-05 | 6.132E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.185E-05 | 8.386E-03 |
|---|
| cellular response to starvation | GO:0009267 |  | 4.446E-05 | 0.01024345 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.446E-05 | 9.105E-03 |
|---|
| protein maturation by peptide bond cleavage | GO:0051605 |  | 5.486E-05 | 0.01011029 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 7.635E-05 | 0.01279233 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.224E-07 | 1.202E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 4.16E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 2.773E-03 |
|---|
| cell fate commitment | GO:0045165 |  | 4.505E-06 | 2.59E-03 |
|---|
| Notch signaling pathway | GO:0007219 |  | 4.902E-06 | 2.255E-03 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 7.197E-06 | 2.759E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.197E-06 | 2.365E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.197E-06 | 2.069E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.255E-06 | 1.854E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.253E-05 | 2.881E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.253E-05 | 2.619E-03 |
|---|



