Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 8996-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2635 | 3.360e-02 | 1.674e-02 | 1.491e-01 |
|---|
| IPC-NIBC-129 | 0.2451 | 7.353e-02 | 6.208e-02 | 3.371e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1885 | 1.312e-02 | 8.379e-03 | 1.579e-01 |
|---|
| Loi_GPL570 | 0.2425 | 2.220e-01 | 2.599e-01 | 6.316e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1376 | 2.180e-04 | 2.600e-05 | 4.210e-04 |
|---|
| Pawitan_GPL96-GPL97 | 0.4356 | 3.064e-01 | 2.282e-01 | 8.533e-01 |
|---|
| Schmidt | 0.1485 | 2.786e-02 | 2.954e-02 | 1.775e-01 |
|---|
| Sotiriou | 0.2634 | 1.719e-01 | 9.050e-02 | 7.445e-01 |
|---|
| Wang | 0.1847 | 6.219e-02 | 4.628e-02 | 2.126e-01 |
|---|
| Zhang | 0.1479 | 1.071e-03 | 4.220e-04 | 7.196e-03 |
|---|
Expression data for subnetwork 8996-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
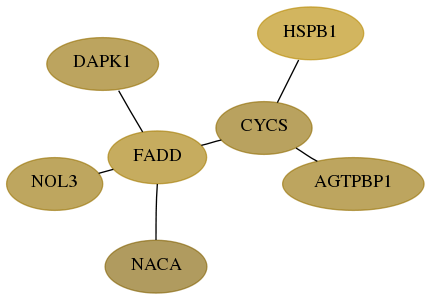
Score for each gene in subnetwork 8996-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| hspb1 |   | 2 | 53 | 115 | 105 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 |
|---|
| agtpbp1 |   | 1 | 90 | 149 | 157 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 |
|---|
| nol3 |   | 1 | 90 | 149 | 157 | 0.089 | 0.110 | 0.102 | -0.119 | 0.094 | 0.125 | -0.023 | 0.016 | 0.180 | 0.059 |
|---|
| cycs |   | 2 | 53 | 115 | 105 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 |
|---|
| dapk1 |   | 2 | 53 | 124 | 113 | 0.082 | 0.188 | -0.078 | -0.063 | 0.067 | 0.267 | 0.117 | 0.059 | -0.061 | 0.142 |
|---|
| fadd |   | 3 | 39 | 1 | 10 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.300 | 0.070 | 0.116 | 0.100 | 0.032 |
|---|
| naca |   | 3 | 39 | 1 | 10 | 0.052 | 0.141 | 0.119 | 0.132 | 0.102 | 0.081 | 0.076 | 0.100 | 0.129 | 0.147 |
|---|
GO Enrichment output for subnetwork 8996-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.236E-06 | 2.844E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 4.06E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.832E-05 | 0.01404755 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.465E-05 | 0.01417505 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.011E-05 | 0.01844835 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.011E-05 | 0.01537363 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.923E-05 | 0.01617412 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 4.923E-05 | 0.01415236 |
|---|
| response to heat | GO:0009408 |  | 1.017E-04 | 0.02599575 |
|---|
| nucleus organization | GO:0006997 |  | 1.234E-04 | 0.02837537 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.007E-04 | 0.04196374 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 5.081E-09 | 1.241E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.153E-08 | 7.516E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.202E-07 | 9.789E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.391E-07 | 8.499E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.828E-07 | 8.93E-05 |
|---|
| nucleus organization | GO:0006997 |  | 8.88E-07 | 3.616E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.551E-06 | 5.412E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.756E-06 | 5.364E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.756E-06 | 4.768E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.617E-06 | 6.392E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 3.057E-06 | 6.79E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.472E-09 | 1.076E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.416E-08 | 6.516E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.058E-07 | 8.487E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.225E-07 | 7.368E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.609E-07 | 7.743E-05 |
|---|
| nucleus organization | GO:0006997 |  | 6.681E-07 | 2.679E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.366E-06 | 4.695E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.366E-06 | 4.108E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.455E-06 | 3.889E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.068E-06 | 4.976E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.56E-06 | 5.599E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 5.081E-09 | 1.241E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.153E-08 | 7.516E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.202E-07 | 9.789E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.391E-07 | 8.499E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.828E-07 | 8.93E-05 |
|---|
| nucleus organization | GO:0006997 |  | 8.88E-07 | 3.616E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.551E-06 | 5.412E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.756E-06 | 5.364E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.756E-06 | 4.768E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.617E-06 | 6.392E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 3.057E-06 | 6.79E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.472E-09 | 1.076E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.416E-08 | 6.516E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.058E-07 | 8.487E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.225E-07 | 7.368E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.609E-07 | 7.743E-05 |
|---|
| nucleus organization | GO:0006997 |  | 6.681E-07 | 2.679E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.366E-06 | 4.695E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.366E-06 | 4.108E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.455E-06 | 3.889E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.068E-06 | 4.976E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.56E-06 | 5.599E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.472E-09 | 1.076E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.416E-08 | 6.516E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.058E-07 | 8.487E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.225E-07 | 7.368E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.609E-07 | 7.743E-05 |
|---|
| nucleus organization | GO:0006997 |  | 6.681E-07 | 2.679E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.366E-06 | 4.695E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.366E-06 | 4.108E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.455E-06 | 3.889E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.068E-06 | 4.976E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.56E-06 | 5.599E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.236E-06 | 2.844E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 4.06E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.832E-05 | 0.01404755 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.465E-05 | 0.01417505 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.011E-05 | 0.01844835 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.011E-05 | 0.01537363 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.923E-05 | 0.01617412 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 4.923E-05 | 0.01415236 |
|---|
| response to heat | GO:0009408 |  | 1.017E-04 | 0.02599575 |
|---|
| nucleus organization | GO:0006997 |  | 1.234E-04 | 0.02837537 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.007E-04 | 0.04196374 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.236E-06 | 2.844E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 4.06E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.832E-05 | 0.01404755 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.465E-05 | 0.01417505 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.011E-05 | 0.01844835 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.011E-05 | 0.01537363 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.923E-05 | 0.01617412 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 4.923E-05 | 0.01415236 |
|---|
| response to heat | GO:0009408 |  | 1.017E-04 | 0.02599575 |
|---|
| nucleus organization | GO:0006997 |  | 1.234E-04 | 0.02837537 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.007E-04 | 0.04196374 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.236E-06 | 2.844E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 4.06E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.832E-05 | 0.01404755 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.465E-05 | 0.01417505 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.011E-05 | 0.01844835 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.011E-05 | 0.01537363 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.923E-05 | 0.01617412 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 4.923E-05 | 0.01415236 |
|---|
| response to heat | GO:0009408 |  | 1.017E-04 | 0.02599575 |
|---|
| nucleus organization | GO:0006997 |  | 1.234E-04 | 0.02837537 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.007E-04 | 0.04196374 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.236E-06 | 2.844E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 4.06E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 1.832E-05 | 0.01404755 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.465E-05 | 0.01417505 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.011E-05 | 0.01844835 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.011E-05 | 0.01537363 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.923E-05 | 0.01617412 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 4.923E-05 | 0.01415236 |
|---|
| response to heat | GO:0009408 |  | 1.017E-04 | 0.02599575 |
|---|
| nucleus organization | GO:0006997 |  | 1.234E-04 | 0.02837537 |
|---|
| cellular component disassembly | GO:0022411 |  | 2.007E-04 | 0.04196374 |
|---|



