Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 80145-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1547 | 2.195e-02 | 9.425e-03 | 1.079e-01 |
|---|
| IPC-NIBC-129 | 0.2127 | 1.288e-01 | 1.169e-01 | 5.203e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2091 | 2.497e-02 | 1.782e-02 | 2.669e-01 |
|---|
| Loi_GPL570 | 0.1975 | 9.729e-02 | 1.222e-01 | 3.719e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1729 | 1.859e-02 | 1.124e-02 | 2.567e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3462 | 1.001e-01 | 5.447e-02 | 4.960e-01 |
|---|
| Schmidt | 0.2126 | 3.112e-02 | 3.291e-02 | 1.944e-01 |
|---|
| Sotiriou | 0.2564 | 9.844e-02 | 4.092e-02 | 5.629e-01 |
|---|
| Wang | 0.2094 | 4.060e-02 | 2.685e-02 | 1.535e-01 |
|---|
| Zhang | 0.2548 | 4.928e-03 | 2.310e-03 | 2.608e-02 |
|---|
Expression data for subnetwork 80145-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
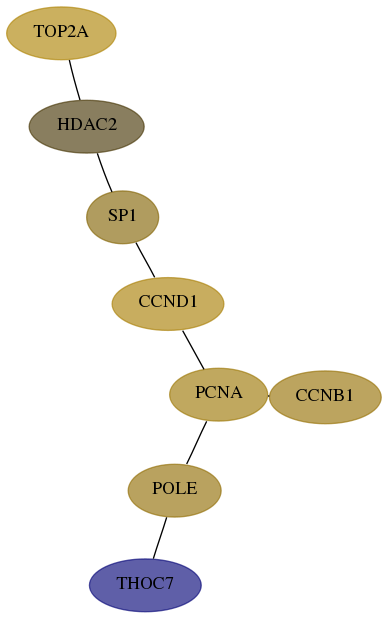
Score for each gene in subnetwork 80145-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| hdac2 |   | 16 | 10 | 64 | 39 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 |
|---|
| pole |   | 3 | 39 | 171 | 151 | 0.073 | 0.135 | 0.081 | -0.205 | 0.081 | 0.139 | 0.145 | 0.105 | 0.068 | -0.014 |
|---|
| top2a |   | 22 | 7 | 70 | 46 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 |
|---|
| ccnb1 |   | 33 | 4 | 68 | 40 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 |
|---|
| thoc7 |   | 1 | 90 | 171 | 178 | -0.040 | 0.148 | 0.110 | -0.113 | 0.072 | 0.135 | -0.066 | 0.095 | 0.134 | 0.219 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 80145-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.754E-07 | 2.018E-04 |
|---|
| interphase | GO:0051325 |  | 1.925E-07 | 1.476E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.256E-06 | 1.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 1.192E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.596E-06 | 1.378E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.596E-06 | 1.181E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 1.158E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 5.323E-06 | 1.36E-03 |
|---|
| DNA topological change | GO:0006265 |  | 5.392E-06 | 1.24E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 1.578E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.069E-08 | 1.238E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.258E-07 | 1.536E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.362E-06 | 1.109E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.551E-06 | 9.472E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.151E-06 | 1.051E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.151E-06 | 8.759E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.617E-06 | 9.132E-04 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.226E-06 | 9.851E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.226E-06 | 8.757E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 4.271E-06 | 1.043E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 6.007E-06 | 1.334E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.86E-08 | 1.65E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.642E-07 | 1.976E-04 |
|---|
| interphase | GO:0051325 |  | 1.862E-07 | 1.493E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.841E-06 | 1.108E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.097E-06 | 1.009E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.644E-06 | 1.06E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.644E-06 | 9.086E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.644E-06 | 7.951E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.352E-06 | 8.961E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.964E-06 | 9.538E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 5.772E-06 | 1.262E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5.069E-08 | 1.238E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.258E-07 | 1.536E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.362E-06 | 1.109E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.551E-06 | 9.472E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.151E-06 | 1.051E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.151E-06 | 8.759E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.617E-06 | 9.132E-04 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.226E-06 | 9.851E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.226E-06 | 8.757E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 4.271E-06 | 1.043E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 6.007E-06 | 1.334E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.86E-08 | 1.65E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.642E-07 | 1.976E-04 |
|---|
| interphase | GO:0051325 |  | 1.862E-07 | 1.493E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.841E-06 | 1.108E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.097E-06 | 1.009E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.644E-06 | 1.06E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.644E-06 | 9.086E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.644E-06 | 7.951E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.352E-06 | 8.961E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.964E-06 | 9.538E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 5.772E-06 | 1.262E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 6.86E-08 | 1.65E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.642E-07 | 1.976E-04 |
|---|
| interphase | GO:0051325 |  | 1.862E-07 | 1.493E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.841E-06 | 1.108E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.097E-06 | 1.009E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.644E-06 | 1.06E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.644E-06 | 9.086E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.644E-06 | 7.951E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.352E-06 | 8.961E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.964E-06 | 9.538E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 5.772E-06 | 1.262E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.754E-07 | 2.018E-04 |
|---|
| interphase | GO:0051325 |  | 1.925E-07 | 1.476E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.256E-06 | 1.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 1.192E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.596E-06 | 1.378E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.596E-06 | 1.181E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 1.158E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 5.323E-06 | 1.36E-03 |
|---|
| DNA topological change | GO:0006265 |  | 5.392E-06 | 1.24E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 1.578E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.754E-07 | 2.018E-04 |
|---|
| interphase | GO:0051325 |  | 1.925E-07 | 1.476E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.256E-06 | 1.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 1.192E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.596E-06 | 1.378E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.596E-06 | 1.181E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 1.158E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 5.323E-06 | 1.36E-03 |
|---|
| DNA topological change | GO:0006265 |  | 5.392E-06 | 1.24E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 1.578E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.754E-07 | 2.018E-04 |
|---|
| interphase | GO:0051325 |  | 1.925E-07 | 1.476E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.256E-06 | 1.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 1.192E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.596E-06 | 1.378E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.596E-06 | 1.181E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 1.158E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 5.323E-06 | 1.36E-03 |
|---|
| DNA topological change | GO:0006265 |  | 5.392E-06 | 1.24E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 1.578E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.033E-07 | 2.376E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.754E-07 | 2.018E-04 |
|---|
| interphase | GO:0051325 |  | 1.925E-07 | 1.476E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.256E-06 | 1.297E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.592E-06 | 1.192E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.596E-06 | 1.378E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.596E-06 | 1.181E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.027E-06 | 1.158E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 5.323E-06 | 1.36E-03 |
|---|
| DNA topological change | GO:0006265 |  | 5.392E-06 | 1.24E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.546E-06 | 1.578E-03 |
|---|



