Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7422-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2607 | 1.112e-02 | 3.738e-03 | 6.338e-02 |
|---|
| IPC-NIBC-129 | 0.2874 | 1.663e-02 | 1.134e-02 | 7.844e-02 |
|---|
| Ivshina_GPL96-GPL97 | 0.2417 | 5.610e-03 | 3.074e-03 | 7.282e-02 |
|---|
| Loi_GPL570 | 0.3585 | 9.371e-02 | 1.181e-01 | 3.621e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1404 | 2.460e-04 | 3.000e-05 | 5.220e-04 |
|---|
| Pawitan_GPL96-GPL97 | 0.3078 | 2.066e-01 | 1.379e-01 | 7.324e-01 |
|---|
| Schmidt | 0.2154 | 1.121e-02 | 1.214e-02 | 8.049e-02 |
|---|
| Sotiriou | 0.3206 | 1.644e-01 | 8.495e-02 | 7.303e-01 |
|---|
| Wang | 0.0937 | 2.835e-01 | 2.979e-01 | 6.022e-01 |
|---|
| Zhang | 0.1866 | 9.451e-03 | 4.780e-03 | 4.463e-02 |
|---|
Expression data for subnetwork 7422-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
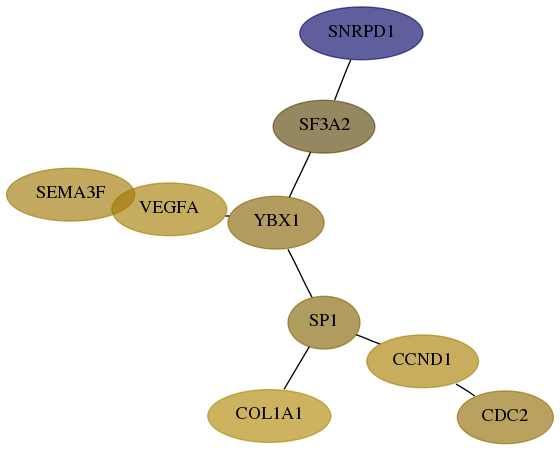
Score for each gene in subnetwork 7422-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| sf3a2 |   | 1 | 90 | 56 | 64 | 0.019 | 0.073 | 0.116 | 0.118 | 0.089 | 0.271 | 0.102 | 0.137 | -0.072 | 0.072 |
|---|
| vegfa |   | 2 | 53 | 56 | 51 | 0.122 | 0.407 | 0.031 | 0.140 | 0.118 | -0.021 | 0.075 | 0.051 | 0.135 | 0.088 |
|---|
| col1a1 |   | 1 | 90 | 56 | 64 | 0.157 | 0.030 | -0.010 | -0.046 | -0.012 | -0.059 | -0.107 | -0.056 | 0.070 | -0.044 |
|---|
| ybx1 |   | 2 | 53 | 56 | 51 | 0.056 | 0.247 | 0.058 | 0.045 | 0.065 | 0.123 | 0.128 | 0.056 | -0.081 | -0.021 |
|---|
| snrpd1 |   | 1 | 90 | 56 | 64 | -0.026 | 0.257 | 0.128 | 0.254 | 0.106 | 0.155 | 0.190 | 0.111 | 0.076 | 0.011 |
|---|
| sema3f |   | 2 | 53 | 56 | 51 | 0.102 | -0.053 | 0.093 | 0.151 | 0.123 | 0.130 | 0.090 | 0.121 | 0.035 | 0.047 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 7422-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 6.87E-06 | 0.01580027 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 0.01156371 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.216E-05 | 0.01273914 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 2.261E-05 | 8.666E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 7.793E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 9.093E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 3.163E-05 | 8.082E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.214E-05 | 9.691E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 4.214E-05 | 8.81E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 1.596E-06 | 3.9E-03 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 4.364E-06 | 5.331E-03 |
|---|
| nuclear mRNA 3'-splice site recognition | GO:0000389 |  | 4.364E-06 | 3.554E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.364E-06 | 2.665E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 4.364E-06 | 2.132E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.585E-06 | 1.867E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 5.849E-06 | 2.041E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.543E-06 | 1.998E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 9.157E-06 | 2.486E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 2.981E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 1.22E-05 | 2.71E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 1.902E-06 | 4.576E-03 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 5.571E-06 | 6.702E-03 |
|---|
| nuclear mRNA 3'-splice site recognition | GO:0000389 |  | 5.571E-06 | 4.468E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.571E-06 | 3.351E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.571E-06 | 2.681E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.579E-06 | 2.638E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.005E-06 | 2.408E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.353E-06 | 2.512E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 1.169E-05 | 3.125E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 3.748E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 1.558E-05 | 3.407E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 1.596E-06 | 3.9E-03 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 4.364E-06 | 5.331E-03 |
|---|
| nuclear mRNA 3'-splice site recognition | GO:0000389 |  | 4.364E-06 | 3.554E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.364E-06 | 2.665E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 4.364E-06 | 2.132E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.585E-06 | 1.867E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 5.849E-06 | 2.041E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.543E-06 | 1.998E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 9.157E-06 | 2.486E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 2.981E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 1.22E-05 | 2.71E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 1.902E-06 | 4.576E-03 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 5.571E-06 | 6.702E-03 |
|---|
| nuclear mRNA 3'-splice site recognition | GO:0000389 |  | 5.571E-06 | 4.468E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.571E-06 | 3.351E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.571E-06 | 2.681E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.579E-06 | 2.638E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.005E-06 | 2.408E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.353E-06 | 2.512E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 1.169E-05 | 3.125E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 3.748E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 1.558E-05 | 3.407E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 1.902E-06 | 4.576E-03 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 5.571E-06 | 6.702E-03 |
|---|
| nuclear mRNA 3'-splice site recognition | GO:0000389 |  | 5.571E-06 | 4.468E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.571E-06 | 3.351E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.571E-06 | 2.681E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.579E-06 | 2.638E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.005E-06 | 2.408E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.353E-06 | 2.512E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 1.169E-05 | 3.125E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 3.748E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 1.558E-05 | 3.407E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 6.87E-06 | 0.01580027 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 0.01156371 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.216E-05 | 0.01273914 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 2.261E-05 | 8.666E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 7.793E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 9.093E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 3.163E-05 | 8.082E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.214E-05 | 9.691E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 4.214E-05 | 8.81E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 6.87E-06 | 0.01580027 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 0.01156371 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.216E-05 | 0.01273914 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 2.261E-05 | 8.666E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 7.793E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 9.093E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 3.163E-05 | 8.082E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.214E-05 | 9.691E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 4.214E-05 | 8.81E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 6.87E-06 | 0.01580027 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 0.01156371 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.216E-05 | 0.01273914 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 2.261E-05 | 8.666E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 7.793E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 9.093E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 3.163E-05 | 8.082E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.214E-05 | 9.691E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 4.214E-05 | 8.81E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosome assembly | GO:0000245 |  | 6.87E-06 | 0.01580027 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 0.01156371 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.216E-05 | 0.01273914 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 2.261E-05 | 8.666E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 7.793E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 9.093E-03 |
|---|
| T-helper 1 type immune response | GO:0042088 |  | 3.163E-05 | 8.082E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.214E-05 | 9.691E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 4.214E-05 | 8.81E-03 |
|---|



