Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 59341-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1695 | 1.224e-02 | 4.261e-03 | 6.839e-02 |
|---|
| IPC-NIBC-129 | 0.2050 | 6.941e-02 | 5.814e-02 | 3.209e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2371 | 2.906e-02 | 2.128e-02 | 2.993e-01 |
|---|
| Loi_GPL570 | 0.2471 | 1.508e-01 | 1.825e-01 | 5.005e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1933 | 1.049e-02 | 5.241e-03 | 1.405e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2826 | 6.750e-02 | 3.288e-02 | 3.829e-01 |
|---|
| Schmidt | 0.1789 | 1.107e-02 | 1.198e-02 | 7.952e-02 |
|---|
| Sotiriou | 0.3215 | 7.087e-02 | 2.560e-02 | 4.605e-01 |
|---|
| Wang | 0.1739 | 7.473e-02 | 5.839e-02 | 2.438e-01 |
|---|
| Zhang | 0.2909 | 1.443e-02 | 7.672e-03 | 6.295e-02 |
|---|
Expression data for subnetwork 59341-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
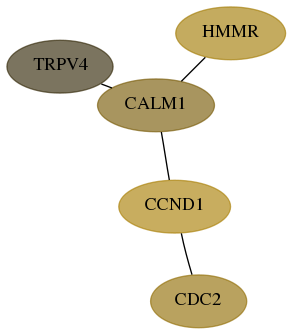
Score for each gene in subnetwork 59341-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| calm1 |   | 16 | 10 | 35 | 19 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| hmmr |   | 10 | 14 | 35 | 23 | 0.112 | 0.310 | 0.147 | 0.127 | 0.145 | 0.221 | 0.179 | 0.233 | 0.227 | 0.144 |
|---|
| trpv4 |   | 1 | 90 | 114 | 120 | 0.006 | 0.209 | 0.101 | 0.051 | 0.071 | 0.072 | -0.001 | 0.127 | -0.005 | -0.067 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 59341-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cell volume homeostasis | GO:0006884 |  | 2.137E-05 | 0.02457665 |
|---|
| response to osmotic stress | GO:0006970 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.05703143 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.05720536 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.05346626 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 2.733E-04 | 0.07856707 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.11666519 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.12138942 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.12087549 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 0.01653866 |
|---|
| cell volume homeostasis | GO:0006884 |  | 1.087E-05 | 0.01328202 |
|---|
| response to osmotic stress | GO:0006970 |  | 2.894E-05 | 0.02357001 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.577E-05 | 0.02795581 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.815E-05 | 0.0381865 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.085E-05 | 0.03698933 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.755E-05 | 0.03404345 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.191E-04 | 0.03635854 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.474E-04 | 0.06715659 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.695E-04 | 0.06583235 |
|---|
| response to calcium ion | GO:0051592 |  | 2.809E-04 | 0.06237559 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| cell volume homeostasis | GO:0006884 |  | 1.188E-05 | 0.01428905 |
|---|
| response to osmotic stress | GO:0006970 |  | 3.162E-05 | 0.02535557 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.02707358 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.03792743 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.03695633 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.03410297 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.3E-04 | 0.03910676 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.702E-04 | 0.07222279 |
|---|
| response to calcium ion | GO:0051592 |  | 2.821E-04 | 0.06786839 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.943E-04 | 0.06436114 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 0.01653866 |
|---|
| cell volume homeostasis | GO:0006884 |  | 1.087E-05 | 0.01328202 |
|---|
| response to osmotic stress | GO:0006970 |  | 2.894E-05 | 0.02357001 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.577E-05 | 0.02795581 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.815E-05 | 0.0381865 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.085E-05 | 0.03698933 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.755E-05 | 0.03404345 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.191E-04 | 0.03635854 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.474E-04 | 0.06715659 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.695E-04 | 0.06583235 |
|---|
| response to calcium ion | GO:0051592 |  | 2.809E-04 | 0.06237559 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| cell volume homeostasis | GO:0006884 |  | 1.188E-05 | 0.01428905 |
|---|
| response to osmotic stress | GO:0006970 |  | 3.162E-05 | 0.02535557 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.02707358 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.03792743 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.03695633 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.03410297 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.3E-04 | 0.03910676 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.702E-04 | 0.07222279 |
|---|
| response to calcium ion | GO:0051592 |  | 2.821E-04 | 0.06786839 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.943E-04 | 0.06436114 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 0.01779296 |
|---|
| cell volume homeostasis | GO:0006884 |  | 1.188E-05 | 0.01428905 |
|---|
| response to osmotic stress | GO:0006970 |  | 3.162E-05 | 0.02535557 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.501E-05 | 0.02707358 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.882E-05 | 0.03792743 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.216E-05 | 0.03695633 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.922E-05 | 0.03410297 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.3E-04 | 0.03910676 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.702E-04 | 0.07222279 |
|---|
| response to calcium ion | GO:0051592 |  | 2.821E-04 | 0.06786839 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.943E-04 | 0.06436114 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cell volume homeostasis | GO:0006884 |  | 2.137E-05 | 0.02457665 |
|---|
| response to osmotic stress | GO:0006970 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.05703143 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.05720536 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.05346626 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 2.733E-04 | 0.07856707 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.11666519 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.12138942 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.12087549 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cell volume homeostasis | GO:0006884 |  | 2.137E-05 | 0.02457665 |
|---|
| response to osmotic stress | GO:0006970 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.05703143 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.05720536 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.05346626 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 2.733E-04 | 0.07856707 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.11666519 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.12138942 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.12087549 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cell volume homeostasis | GO:0006884 |  | 2.137E-05 | 0.02457665 |
|---|
| response to osmotic stress | GO:0006970 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.05703143 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.05720536 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.05346626 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 2.733E-04 | 0.07856707 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.11666519 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.12138942 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.12087549 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.248E-05 | 0.02869902 |
|---|
| cell volume homeostasis | GO:0006884 |  | 2.137E-05 | 0.02457665 |
|---|
| response to osmotic stress | GO:0006970 |  | 6.216E-05 | 0.04765688 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 7.101E-05 | 0.04083006 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.24E-04 | 0.05703143 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.492E-04 | 0.05720536 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.627E-04 | 0.05346626 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 2.733E-04 | 0.07856707 |
|---|
| response to calcium ion | GO:0051592 |  | 4.565E-04 | 0.11666519 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.278E-04 | 0.12138942 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.781E-04 | 0.12087549 |
|---|



