Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2017-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1522 | 4.579e-03 | 1.107e-03 | 3.099e-02 |
|---|
| IPC-NIBC-129 | 0.1713 | 4.242e-02 | 3.318e-02 | 2.053e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2838 | 5.624e-02 | 4.585e-02 | 4.702e-01 |
|---|
| Loi_GPL570 | 0.2859 | 8.771e-02 | 1.111e-01 | 3.452e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1743 | 2.047e-02 | 1.276e-02 | 2.815e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3948 | 4.686e-02 | 2.059e-02 | 2.942e-01 |
|---|
| Schmidt | 0.2204 | 2.574e-02 | 2.734e-02 | 1.662e-01 |
|---|
| Sotiriou | 0.2684 | 9.629e-02 | 3.965e-02 | 5.558e-01 |
|---|
| Wang | 0.1810 | 6.621e-02 | 5.011e-02 | 2.228e-01 |
|---|
| Zhang | 0.3240 | 2.156e-03 | 9.190e-04 | 1.304e-02 |
|---|
Expression data for subnetwork 2017-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
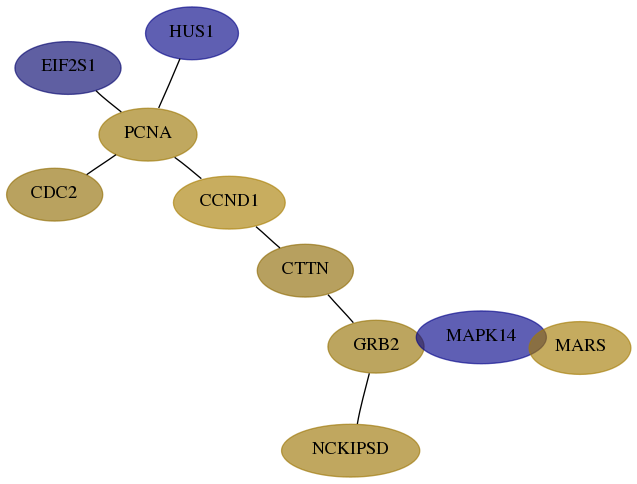
Score for each gene in subnetwork 2017-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| cttn |   | 5 | 20 | 8 | 9 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.216 | 0.069 | 0.115 | 0.129 | 0.025 |
|---|
| hus1 |   | 6 | 17 | 22 | 17 | -0.058 | 0.197 | 0.116 | 0.109 | 0.101 | 0.160 | 0.072 | 0.082 | 0.081 | 0.071 |
|---|
| mapk14 |   | 2 | 53 | 22 | 31 | -0.062 | 0.058 | 0.135 | 0.204 | 0.086 | 0.068 | 0.246 | 0.068 | 0.056 | 0.011 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| nckipsd |   | 1 | 90 | 22 | 41 | 0.094 | 0.086 | 0.221 | -0.049 | 0.109 | 0.121 | 0.006 | 0.126 | 0.073 | 0.054 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| mars |   | 2 | 53 | 22 | 31 | 0.114 | 0.207 | 0.114 | 0.083 | 0.198 | 0.340 | 0.139 | 0.180 | 0.050 | 0.129 |
|---|
| eif2s1 |   | 13 | 12 | 8 | 4 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 |
|---|
| grb2 |   | 6 | 17 | 8 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 |
|---|
GO Enrichment output for subnetwork 2017-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.429E-06 | 3.287E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 0.04330314 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.105E-05 | 0.03147128 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.282E-05 | 0.02462315 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.071E-04 | 0.04928164 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.566E-04 | 0.06003288 |
|---|
| nuclear import | GO:0051170 |  | 1.63E-04 | 0.05355026 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 1.768E-04 | 0.05082779 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.974E-04 | 0.05043881 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.327E-04 | 0.05352658 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.327E-04 | 0.04866053 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.174E-07 | 5.312E-04 |
|---|
| response to UV | GO:0009411 |  | 9.988E-06 | 0.01219979 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.055E-05 | 8.589E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.552E-05 | 9.476E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 3.326E-05 | 0.01624924 |
|---|
| nuclear import | GO:0051170 |  | 3.449E-05 | 0.01404172 |
|---|
| neuropeptide signaling pathway | GO:0007218 |  | 3.97E-05 | 0.01385651 |
|---|
| protein localization in nucleus | GO:0034504 |  | 4.25E-05 | 0.0129771 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 4.312E-05 | 0.01170448 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 5.495E-05 | 0.01342406 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 5.799E-05 | 0.01287921 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 3.57E-07 | 8.59E-04 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 0.01757397 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.726E-05 | 0.0138387 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 0.0129921 |
|---|
| neuropeptide signaling pathway | GO:0007218 |  | 4.497E-05 | 0.02163811 |
|---|
| protein import into nucleus | GO:0006606 |  | 5.042E-05 | 0.0202165 |
|---|
| nuclear import | GO:0051170 |  | 5.232E-05 | 0.01798416 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 6E-05 | 0.01804614 |
|---|
| protein localization in nucleus | GO:0034504 |  | 6.477E-05 | 0.01731506 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 8.068E-05 | 0.01941281 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.956E-05 | 0.01958891 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.174E-07 | 5.312E-04 |
|---|
| response to UV | GO:0009411 |  | 9.988E-06 | 0.01219979 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.055E-05 | 8.589E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.552E-05 | 9.476E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 3.326E-05 | 0.01624924 |
|---|
| nuclear import | GO:0051170 |  | 3.449E-05 | 0.01404172 |
|---|
| neuropeptide signaling pathway | GO:0007218 |  | 3.97E-05 | 0.01385651 |
|---|
| protein localization in nucleus | GO:0034504 |  | 4.25E-05 | 0.0129771 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 4.312E-05 | 0.01170448 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 5.495E-05 | 0.01342406 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 5.799E-05 | 0.01287921 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 3.57E-07 | 8.59E-04 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 0.01757397 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.726E-05 | 0.0138387 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 0.0129921 |
|---|
| neuropeptide signaling pathway | GO:0007218 |  | 4.497E-05 | 0.02163811 |
|---|
| protein import into nucleus | GO:0006606 |  | 5.042E-05 | 0.0202165 |
|---|
| nuclear import | GO:0051170 |  | 5.232E-05 | 0.01798416 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 6E-05 | 0.01804614 |
|---|
| protein localization in nucleus | GO:0034504 |  | 6.477E-05 | 0.01731506 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 8.068E-05 | 0.01941281 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.956E-05 | 0.01958891 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 3.57E-07 | 8.59E-04 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 0.01757397 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.726E-05 | 0.0138387 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 0.0129921 |
|---|
| neuropeptide signaling pathway | GO:0007218 |  | 4.497E-05 | 0.02163811 |
|---|
| protein import into nucleus | GO:0006606 |  | 5.042E-05 | 0.0202165 |
|---|
| nuclear import | GO:0051170 |  | 5.232E-05 | 0.01798416 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 6E-05 | 0.01804614 |
|---|
| protein localization in nucleus | GO:0034504 |  | 6.477E-05 | 0.01731506 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 8.068E-05 | 0.01941281 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.956E-05 | 0.01958891 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.429E-06 | 3.287E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 0.04330314 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.105E-05 | 0.03147128 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.282E-05 | 0.02462315 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.071E-04 | 0.04928164 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.566E-04 | 0.06003288 |
|---|
| nuclear import | GO:0051170 |  | 1.63E-04 | 0.05355026 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 1.768E-04 | 0.05082779 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.974E-04 | 0.05043881 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.327E-04 | 0.05352658 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.327E-04 | 0.04866053 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.429E-06 | 3.287E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 0.04330314 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.105E-05 | 0.03147128 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.282E-05 | 0.02462315 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.071E-04 | 0.04928164 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.566E-04 | 0.06003288 |
|---|
| nuclear import | GO:0051170 |  | 1.63E-04 | 0.05355026 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 1.768E-04 | 0.05082779 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.974E-04 | 0.05043881 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.327E-04 | 0.05352658 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.327E-04 | 0.04866053 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.429E-06 | 3.287E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 0.04330314 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.105E-05 | 0.03147128 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.282E-05 | 0.02462315 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.071E-04 | 0.04928164 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.566E-04 | 0.06003288 |
|---|
| nuclear import | GO:0051170 |  | 1.63E-04 | 0.05355026 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 1.768E-04 | 0.05082779 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.974E-04 | 0.05043881 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.327E-04 | 0.05352658 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.327E-04 | 0.04866053 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.429E-06 | 3.287E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 0.04330314 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.105E-05 | 0.03147128 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.282E-05 | 0.02462315 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 1.071E-04 | 0.04928164 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.566E-04 | 0.06003288 |
|---|
| nuclear import | GO:0051170 |  | 1.63E-04 | 0.05355026 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 1.768E-04 | 0.05082779 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.974E-04 | 0.05043881 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.327E-04 | 0.05352658 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.327E-04 | 0.04866053 |
|---|



