Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 92359-11-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2148 | 3.172e-02 | 2.759e-02 | 2.099e-01 |
|---|
| IPC-NIBC-129 | 0.2971 | 3.987e-02 | 5.795e-02 | 3.842e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1934 | 3.359e-03 | 6.135e-03 | 1.437e-01 |
|---|
| Loi_GPL570 | 0.3619 | 1.158e-02 | 1.366e-02 | 6.250e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1403 | 0.000e+00 | 2.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2898 | 1.015e-02 | 1.353e-02 | 1.487e-01 |
|---|
| Parker_GPL887 | 0.0981 | 3.282e-01 | 3.467e-01 | 3.532e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3594 | 5.504e-01 | 4.798e-01 | 9.777e-01 |
|---|
| Schmidt | 0.3525 | 1.305e-02 | 1.465e-02 | 1.737e-01 |
|---|
| Sotiriou | 0.3479 | 6.678e-03 | 5.049e-03 | 1.490e-01 |
|---|
| Van-De-Vijver | 0.2734 | 9.570e-04 | 1.377e-03 | 5.700e-02 |
|---|
| Zhang | 0.2966 | 1.802e-01 | 1.274e-01 | 3.565e-01 |
|---|
| Zhou | 0.4228 | 6.682e-02 | 5.523e-02 | 3.930e-01 |
|---|
Expression data for subnetwork 92359-11-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
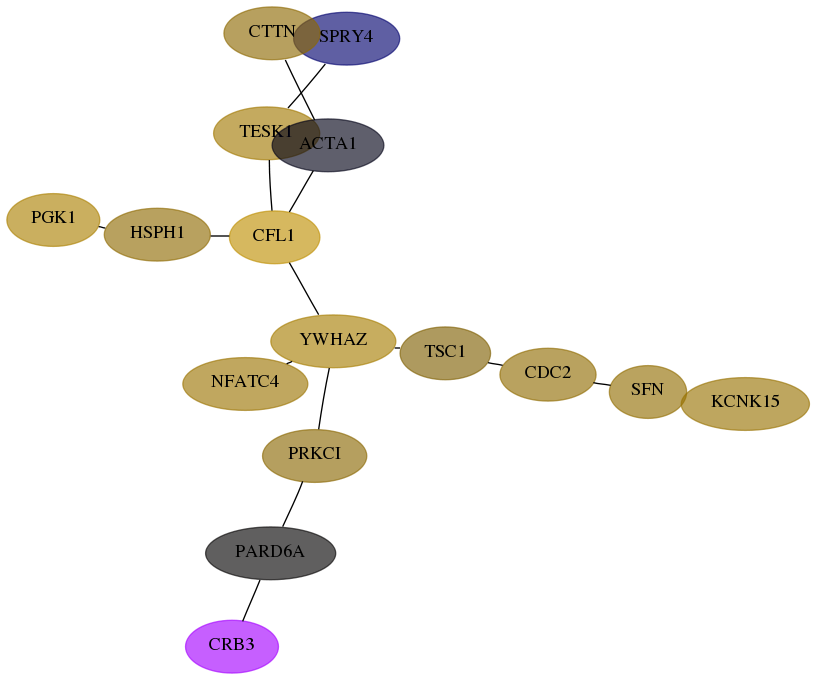
Score for each gene in subnetwork 92359-11-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| kcnk15 |   | 4 | 50 | 1 | 10 | 0.088 | -0.091 | -0.068 | 0.220 | 0.076 | -0.136 | 0.026 | 0.083 | 0.037 | 0.154 | undef | 0.098 | 0.140 |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| cttn |   | 10 | 20 | 1 | 6 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | undef | 0.025 | 0.279 |
|---|
| pard6a |   | 1 | 116 | 31 | 51 | 0.000 | 0.045 | 0.068 | -0.013 | 0.115 | -0.148 | 0.183 | -0.003 | 0.094 | 0.023 | 0.098 | 0.084 | 0.030 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| pgk1 |   | 5 | 40 | 18 | 23 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | -0.046 | 0.054 | 0.141 |
|---|
| cfl1 |   | 8 | 26 | 18 | 16 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.251 | -0.466 | 0.229 | 0.174 | 0.188 | 0.208 | -0.025 | 0.215 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hsph1 |   | 8 | 26 | 18 | 16 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | -0.006 | 0.094 | 0.357 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| acta1 |   | 9 | 22 | 1 | 7 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | 0.100 | 0.048 | 0.201 |
|---|
| tesk1 |   | 7 | 30 | 18 | 19 | 0.110 | 0.169 | 0.161 | 0.179 | 0.096 | 0.138 | -0.154 | 0.245 | 0.155 | 0.092 | 0.067 | 0.029 | 0.200 |
|---|
| spry4 |   | 5 | 40 | 18 | 23 | -0.033 | 0.158 | -0.001 | -0.039 | 0.103 | 0.074 | -0.328 | 0.022 | 0.197 | 0.156 | undef | 0.184 | 0.211 |
|---|
| crb3 |   | 1 | 116 | 31 | 51 | undef | 0.136 | 0.092 | 0.107 | 0.096 | undef | 0.084 | 0.128 | undef | undef | undef | undef | undef |
|---|
GO Enrichment output for subnetwork 92359-11-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.128E-09 | 9.494E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.031E-08 | 1.186E-05 |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 3.488E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.55E-08 | 2.616E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.804E-07 | 1.75E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.804E-07 | 1.458E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 2.449E-04 |
|---|
| glycolysis | GO:0006096 |  | 9.118E-07 | 2.621E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.099E-06 | 5.364E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 5.786E-04 |
|---|
| actin filament organization | GO:0007015 |  | 3.003E-06 | 6.278E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.678E-10 | 1.143E-06 |
|---|
| establishment of cell polarity | GO:0030010 |  | 5.461E-10 | 6.671E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.126E-08 | 9.166E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.126E-08 | 6.875E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 1.126E-08 | 5.5E-06 |
|---|
| actin filament organization | GO:0007015 |  | 1.411E-08 | 5.744E-06 |
|---|
| neural plate development | GO:0001840 |  | 2.25E-08 | 7.851E-06 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.093E-07 | 3.337E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.754E-07 | 4.762E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.849E-07 | 4.517E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.2E-07 | 7.107E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.727E-10 | 1.378E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.312E-08 | 1.579E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.312E-08 | 1.053E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.569E-07 | 9.437E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.951E-07 | 9.39E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.156E-07 | 8.644E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.73E-07 | 1.282E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.431E-07 | 1.333E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.288E-07 | 1.948E-04 |
|---|
| actin filament organization | GO:0007015 |  | 9.282E-07 | 2.233E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 9.864E-07 | 2.157E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.678E-10 | 1.143E-06 |
|---|
| establishment of cell polarity | GO:0030010 |  | 5.461E-10 | 6.671E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.126E-08 | 9.166E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.126E-08 | 6.875E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 1.126E-08 | 5.5E-06 |
|---|
| actin filament organization | GO:0007015 |  | 1.411E-08 | 5.744E-06 |
|---|
| neural plate development | GO:0001840 |  | 2.25E-08 | 7.851E-06 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.093E-07 | 3.337E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.754E-07 | 4.762E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.849E-07 | 4.517E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.2E-07 | 7.107E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.727E-10 | 1.378E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.312E-08 | 1.579E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.312E-08 | 1.053E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.569E-07 | 9.437E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.951E-07 | 9.39E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.156E-07 | 8.644E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.73E-07 | 1.282E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.431E-07 | 1.333E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.288E-07 | 1.948E-04 |
|---|
| actin filament organization | GO:0007015 |  | 9.282E-07 | 2.233E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 9.864E-07 | 2.157E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell cortex | GO:0005938 |  | 1.544E-11 | 5.514E-08 |
|---|
| establishment of cell polarity | GO:0030010 |  | 6.416E-08 | 1.146E-04 |
|---|
| actin filament organization | GO:0007015 |  | 1.304E-07 | 1.553E-04 |
|---|
| cell-cell junction organization | GO:0045216 |  | 3.623E-07 | 3.234E-04 |
|---|
| actin filament | GO:0005884 |  | 1.22E-06 | 8.716E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 2.883E-06 | 1.716E-03 |
|---|
| cell junction organization | GO:0034330 |  | 2.883E-06 | 1.471E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 7.176E-06 | 3.203E-03 |
|---|
| neural plate development | GO:0001840 |  | 7.176E-06 | 2.847E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.176E-06 | 2.562E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 7.176E-06 | 2.329E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell cortex | GO:0005938 |  | 2.674E-09 | 9.756E-06 |
|---|
| establishment of cell polarity | GO:0030010 |  | 4.29E-08 | 7.824E-05 |
|---|
| actin filament organization | GO:0007015 |  | 1.574E-07 | 1.914E-04 |
|---|
| cell-cell junction organization | GO:0045216 |  | 6.749E-07 | 6.156E-04 |
|---|
| actin filament | GO:0005884 |  | 1.026E-06 | 7.482E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.847E-06 | 1.123E-03 |
|---|
| cell junction organization | GO:0034330 |  | 3.91E-06 | 2.038E-03 |
|---|
| lamellipodium | GO:0030027 |  | 5.354E-06 | 2.441E-03 |
|---|
| muscle thin filament assembly | GO:0030240 |  | 6.839E-06 | 2.772E-03 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 6.839E-06 | 2.495E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 6.839E-06 | 2.268E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.727E-10 | 1.378E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.312E-08 | 1.579E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.312E-08 | 1.053E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.569E-07 | 9.437E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.951E-07 | 9.39E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.156E-07 | 8.644E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.73E-07 | 1.282E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.431E-07 | 1.333E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.288E-07 | 1.948E-04 |
|---|
| actin filament organization | GO:0007015 |  | 9.282E-07 | 2.233E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 9.864E-07 | 2.157E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.128E-09 | 9.494E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.031E-08 | 1.186E-05 |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 3.488E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.55E-08 | 2.616E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.804E-07 | 1.75E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.804E-07 | 1.458E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 2.449E-04 |
|---|
| glycolysis | GO:0006096 |  | 9.118E-07 | 2.621E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.099E-06 | 5.364E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 5.786E-04 |
|---|
| actin filament organization | GO:0007015 |  | 3.003E-06 | 6.278E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.128E-09 | 9.494E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.031E-08 | 1.186E-05 |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 3.488E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.55E-08 | 2.616E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.804E-07 | 1.75E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.804E-07 | 1.458E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 2.449E-04 |
|---|
| glycolysis | GO:0006096 |  | 9.118E-07 | 2.621E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.099E-06 | 5.364E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 5.786E-04 |
|---|
| actin filament organization | GO:0007015 |  | 3.003E-06 | 6.278E-04 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.036E-08 | 5.595E-05 |
|---|
| actin filament organization | GO:0007015 |  | 6.641E-07 | 6.12E-04 |
|---|
| cell-cell junction organization | GO:0045216 |  | 8.623E-07 | 5.297E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 3.416E-06 | 1.574E-03 |
|---|
| cell junction organization | GO:0034330 |  | 4.607E-06 | 1.698E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.318E-05 | 7.121E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 2.318E-05 | 6.103E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 3.474E-05 | 8.004E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.474E-05 | 7.114E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 3.474E-05 | 6.403E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.474E-05 | 5.821E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.128E-09 | 9.494E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.031E-08 | 1.186E-05 |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 3.488E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.55E-08 | 2.616E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.804E-07 | 1.75E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.804E-07 | 1.458E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 2.449E-04 |
|---|
| glycolysis | GO:0006096 |  | 9.118E-07 | 2.621E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.099E-06 | 5.364E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 5.786E-04 |
|---|
| actin filament organization | GO:0007015 |  | 3.003E-06 | 6.278E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 4.535E-09 | 1.076E-05 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.132E-08 | 1.343E-05 |
|---|
| polyol catabolic process | GO:0046174 |  | 3.776E-08 | 2.986E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.776E-08 | 2.24E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 4.506E-07 | 2.138E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 4.506E-07 | 1.782E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.243E-07 | 2.794E-04 |
|---|
| glycolysis | GO:0006096 |  | 1.111E-06 | 3.297E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.526E-06 | 6.661E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.534E-06 | 6.013E-04 |
|---|
| cell-cell junction organization | GO:0045216 |  | 3.037E-06 | 6.552E-04 |
|---|



