Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 8607-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1509 | 3.920e-02 | 3.497e-02 | 2.440e-01 |
|---|
| IPC-NIBC-129 | 0.3124 | 4.827e-02 | 6.957e-02 | 4.352e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1829 | 6.056e-03 | 1.052e-02 | 2.050e-01 |
|---|
| Loi_GPL570 | 0.3814 | 8.660e-03 | 1.028e-02 | 4.795e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1179 | 0.000e+00 | 1.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2629 | 1.171e-02 | 1.551e-02 | 1.634e-01 |
|---|
| Parker_GPL887 | 0.2278 | 4.829e-01 | 5.019e-01 | 5.426e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3072 | 1.536e-01 | 1.037e-01 | 6.703e-01 |
|---|
| Schmidt | 0.3428 | 1.816e-02 | 2.087e-02 | 2.190e-01 |
|---|
| Sotiriou | 0.2904 | 1.742e-02 | 1.355e-02 | 2.568e-01 |
|---|
| Van-De-Vijver | 0.2595 | 2.152e-03 | 3.092e-03 | 9.693e-02 |
|---|
| Zhang | 0.2766 | 2.500e-01 | 1.918e-01 | 4.598e-01 |
|---|
| Zhou | 0.3774 | 1.072e-01 | 1.014e-01 | 5.425e-01 |
|---|
Expression data for subnetwork 8607-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
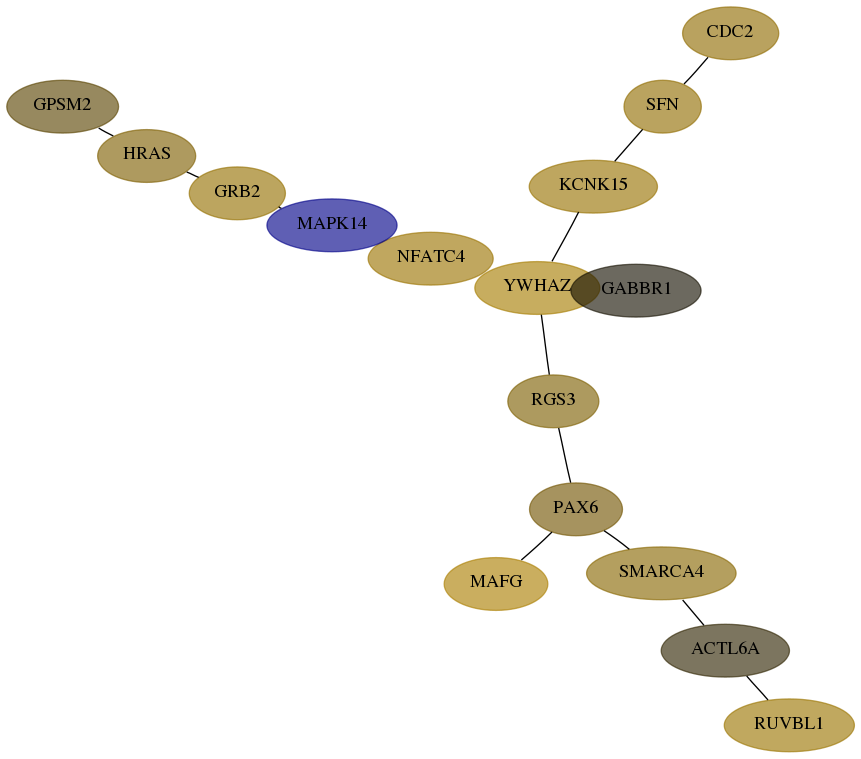
Score for each gene in subnetwork 8607-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| rgs3 |   | 4 | 50 | 77 | 66 | 0.048 | 0.195 | 0.029 | 0.052 | 0.056 | 0.192 | -0.052 | -0.078 | 0.033 | 0.023 | undef | 0.100 | -0.196 |
|---|
| mapk14 |   | 4 | 50 | 1 | 10 | -0.062 | 0.058 | 0.135 | 0.204 | 0.086 | 0.061 | 0.389 | 0.068 | 0.246 | 0.068 | 0.076 | 0.011 | 0.192 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| ruvbl1 |   | 1 | 116 | 90 | 97 | 0.096 | 0.187 | 0.151 | 0.078 | 0.130 | 0.092 | -0.090 | 0.262 | 0.062 | 0.108 | -0.023 | 0.007 | 0.238 |
|---|
| gabbr1 |   | 7 | 30 | 56 | 49 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.125 | 0.096 | 0.116 |
|---|
| kcnk15 |   | 4 | 50 | 1 | 10 | 0.088 | -0.091 | -0.068 | 0.220 | 0.076 | -0.136 | 0.026 | 0.083 | 0.037 | 0.154 | undef | 0.098 | 0.140 |
|---|
| actl6a |   | 1 | 116 | 90 | 97 | 0.007 | 0.244 | 0.085 | 0.212 | 0.091 | 0.106 | 0.191 | 0.114 | 0.270 | 0.105 | undef | 0.213 | 0.250 |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| mafg |   | 1 | 116 | 90 | 97 | 0.136 | 0.178 | -0.061 | 0.150 | 0.069 | -0.150 | 0.267 | 0.007 | 0.233 | 0.017 | 0.125 | -0.071 | 0.289 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| gpsm2 |   | 4 | 50 | 42 | 40 | 0.021 | 0.286 | 0.155 | 0.144 | 0.108 | 0.242 | 0.307 | 0.134 | 0.234 | 0.077 | 0.112 | 0.098 | 0.207 |
|---|
| smarca4 |   | 1 | 116 | 90 | 97 | 0.062 | 0.223 | 0.136 | 0.131 | 0.107 | 0.119 | -0.044 | 0.222 | 0.120 | 0.093 | 0.107 | 0.008 | 0.281 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| pax6 |   | 1 | 116 | 90 | 97 | 0.036 | 0.231 | -0.065 | 0.113 | 0.068 | 0.137 | 0.081 | -0.018 | 0.179 | 0.131 | undef | 0.070 | -0.140 |
|---|
| grb2 |   | 9 | 22 | 1 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | 0.149 | 0.194 | -0.005 |
|---|
GO Enrichment output for subnetwork 8607-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.557E-08 | 1.278E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 9.101E-07 | 1.047E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.07E-06 | 2.353E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.218E-06 | 3.575E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.098E-05 | 5.053E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.971E-05 | 7.555E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.665E-05 | 8.757E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 3.205E-05 | 9.213E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.208E-05 | 8.197E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 3.235E-05 | 7.44E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 3.504E-05 | 7.326E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.646E-08 | 4.022E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.677E-07 | 5.713E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.134E-07 | 7.438E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.16E-06 | 1.319E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.205E-06 | 2.055E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.205E-06 | 1.712E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.29E-06 | 1.846E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.982E-06 | 2.437E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.145E-05 | 3.108E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.245E-05 | 3.042E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.426E-05 | 3.168E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.337E-08 | 5.623E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.634E-07 | 7.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.295E-06 | 1.039E-03 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.627E-06 | 1.58E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.646E-06 | 2.236E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.275E-06 | 2.115E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 5.275E-06 | 1.813E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.13E-05 | 3.398E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.621E-05 | 4.332E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.621E-05 | 3.899E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.803E-05 | 3.945E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.646E-08 | 4.022E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.677E-07 | 5.713E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.134E-07 | 7.438E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.16E-06 | 1.319E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.205E-06 | 2.055E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.205E-06 | 1.712E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.29E-06 | 1.846E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.982E-06 | 2.437E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.145E-05 | 3.108E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.245E-05 | 3.042E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.426E-05 | 3.168E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.337E-08 | 5.623E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.634E-07 | 7.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.295E-06 | 1.039E-03 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.627E-06 | 1.58E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.646E-06 | 2.236E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.275E-06 | 2.115E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 5.275E-06 | 1.813E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.13E-05 | 3.398E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.621E-05 | 4.332E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.621E-05 | 3.899E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.803E-05 | 3.945E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NuA4 histone acetyltransferase complex | GO:0035267 |  | 4.453E-08 | 1.59E-04 |
|---|
| H4/H2A histone acetyltransferase complex | GO:0043189 |  | 7.119E-08 | 1.271E-04 |
|---|
| histone acetyltransferase complex | GO:0000123 |  | 2.789E-07 | 3.32E-04 |
|---|
| SWI/SNF-type complex | GO:0070603 |  | 2.789E-07 | 2.49E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 4.268E-06 | 3.048E-03 |
|---|
| SAP kinase activity | GO:0016909 |  | 1.25E-05 | 7.442E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 1.53E-05 | 7.806E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 1.874E-05 | 8.367E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.874E-05 | 7.437E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.874E-05 | 6.694E-03 |
|---|
| epidermal cell differentiation | GO:0009913 |  | 2.137E-05 | 6.937E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NuA4 histone acetyltransferase complex | GO:0035267 |  | 2.428E-08 | 8.857E-05 |
|---|
| H4/H2A histone acetyltransferase complex | GO:0043189 |  | 4.246E-08 | 7.745E-05 |
|---|
| histone acetyltransferase complex | GO:0000123 |  | 2.659E-07 | 3.234E-04 |
|---|
| SWI/SNF-type complex | GO:0070603 |  | 4.393E-07 | 4.007E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 3.202E-06 | 2.336E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.196E-05 | 7.272E-03 |
|---|
| epidermal growth factor receptor binding | GO:0005154 |  | 1.196E-05 | 6.234E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.793E-05 | 8.176E-03 |
|---|
| SAP kinase activity | GO:0016909 |  | 1.793E-05 | 7.268E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.793E-05 | 6.541E-03 |
|---|
| chromatin remodeling complex | GO:0016585 |  | 2.037E-05 | 6.756E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.337E-08 | 5.623E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.634E-07 | 7.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.295E-06 | 1.039E-03 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.627E-06 | 1.58E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.646E-06 | 2.236E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.275E-06 | 2.115E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 5.275E-06 | 1.813E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.13E-05 | 3.398E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.621E-05 | 4.332E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.621E-05 | 3.899E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 1.803E-05 | 3.945E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.557E-08 | 1.278E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 9.101E-07 | 1.047E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.07E-06 | 2.353E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.218E-06 | 3.575E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.098E-05 | 5.053E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.971E-05 | 7.555E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.665E-05 | 8.757E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 3.205E-05 | 9.213E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.208E-05 | 8.197E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 3.235E-05 | 7.44E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 3.504E-05 | 7.326E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.557E-08 | 1.278E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 9.101E-07 | 1.047E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.07E-06 | 2.353E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.218E-06 | 3.575E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.098E-05 | 5.053E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.971E-05 | 7.555E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.665E-05 | 8.757E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 3.205E-05 | 9.213E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.208E-05 | 8.197E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 3.235E-05 | 7.44E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 3.504E-05 | 7.326E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Ras protein signal transduction | GO:0007265 |  | 2.576E-06 | 4.747E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 9.748E-06 | 8.983E-03 |
|---|
| epidermal cell differentiation | GO:0009913 |  | 1.334E-05 | 8.195E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 2.318E-05 | 0.01068088 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.318E-05 | 8.545E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 3.474E-05 | 0.01067157 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 3.474E-05 | 9.147E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 4.86E-05 | 0.01119537 |
|---|
| visual behavior | GO:0007632 |  | 4.86E-05 | 9.951E-03 |
|---|
| embryonic hindlimb morphogenesis | GO:0035116 |  | 6.474E-05 | 0.01193131 |
|---|
| regulation of neuronal synaptic plasticity | GO:0048168 |  | 6.474E-05 | 0.01084665 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.557E-08 | 1.278E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 9.101E-07 | 1.047E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.07E-06 | 2.353E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.218E-06 | 3.575E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.098E-05 | 5.053E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.971E-05 | 7.555E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.665E-05 | 8.757E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 3.205E-05 | 9.213E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.208E-05 | 8.197E-03 |
|---|
| glial cell fate commitment | GO:0021781 |  | 3.235E-05 | 7.44E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 3.504E-05 | 7.326E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.433E-08 | 1.289E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.185E-06 | 1.406E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.64E-06 | 2.879E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 8.192E-06 | 4.86E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.376E-05 | 6.531E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.364E-05 | 9.352E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.136E-05 | 0.01063106 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 4.057E-05 | 0.01203354 |
|---|
| polyol metabolic process | GO:0019751 |  | 4.057E-05 | 0.01069648 |
|---|
| glial cell fate commitment | GO:0021781 |  | 4.757E-05 | 0.01128803 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 4.757E-05 | 0.01026185 |
|---|



