Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7112-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2035 | 2.111e-02 | 1.745e-02 | 1.555e-01 |
|---|
| IPC-NIBC-129 | 0.3775 | 6.441e-03 | 9.904e-03 | 8.844e-02 |
|---|
| Ivshina_GPL96-GPL97 | 0.2134 | 2.831e-02 | 4.295e-02 | 4.612e-01 |
|---|
| Loi_GPL570 | 0.2878 | 3.415e-02 | 3.924e-02 | 1.602e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1762 | 0.000e+00 | 0.000e+00 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.1914 | 1.009e-01 | 1.210e-01 | 5.630e-01 |
|---|
| Parker_GPL887 | 0.0431 | 3.520e-01 | 3.707e-01 | 3.826e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3575 | 8.386e-01 | 8.028e-01 | 9.994e-01 |
|---|
| Schmidt | 0.1935 | 3.334e-02 | 3.982e-02 | 3.263e-01 |
|---|
| Sotiriou | 0.3034 | 1.405e-02 | 1.086e-02 | 2.282e-01 |
|---|
| Van-De-Vijver | 0.4037 | 7.230e-04 | 1.040e-03 | 4.714e-02 |
|---|
| Zhang | 0.2395 | 1.052e-01 | 6.477e-02 | 2.290e-01 |
|---|
| Zhou | 0.4385 | 6.802e-02 | 5.653e-02 | 3.982e-01 |
|---|
Expression data for subnetwork 7112-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
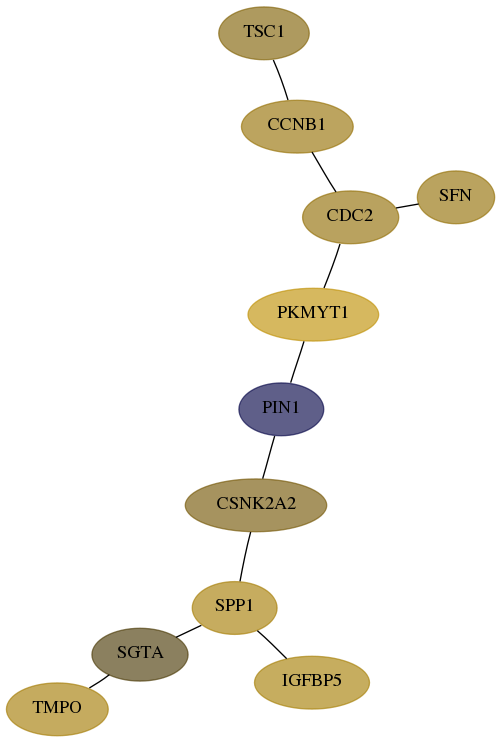
Score for each gene in subnetwork 7112-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| pin1 |   | 1 | 116 | 112 | 123 | -0.012 | 0.181 | 0.133 | 0.270 | 0.112 | 0.074 | -0.085 | 0.072 | 0.063 | 0.161 | 0.057 | 0.259 | 0.019 |
|---|
| tmpo |   | 2 | 78 | 112 | 109 | 0.116 | 0.177 | 0.104 | 0.256 | 0.163 | 0.152 | 0.068 | 0.284 | 0.099 | 0.144 | 0.148 | 0.130 | 0.309 |
|---|
| spp1 |   | 3 | 62 | 106 | 95 | 0.121 | 0.316 | 0.018 | -0.074 | 0.132 | 0.124 | -0.237 | 0.185 | 0.031 | 0.109 | 0.212 | 0.141 | 0.275 |
|---|
| csnk2a2 |   | 2 | 78 | 106 | 103 | 0.036 | 0.182 | 0.089 | -0.033 | 0.074 | 0.094 | -0.125 | 0.149 | 0.060 | 0.090 | 0.051 | -0.155 | 0.174 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| igfbp5 |   | 2 | 78 | 112 | 109 | 0.123 | 0.143 | 0.025 | 0.150 | 0.143 | 0.006 | -0.178 | -0.055 | 0.063 | 0.147 | 0.238 | 0.026 | -0.043 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| pkmyt1 |   | 1 | 116 | 112 | 123 | 0.219 | 0.222 | 0.151 | 0.190 | 0.092 | undef | 0.460 | 0.202 | 0.156 | 0.061 | 0.326 | 0.288 | 0.257 |
|---|
| sgta |   | 2 | 78 | 112 | 109 | 0.013 | 0.164 | 0.126 | 0.061 | 0.074 | 0.070 | 0.298 | 0.111 | 0.036 | 0.035 | 0.014 | 0.044 | 0.129 |
|---|
GO Enrichment output for subnetwork 7112-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 6.041E-07 | 1.389E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.01284463 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.193E-05 | 0.01260993 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 2.343E-05 | 0.0107755 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 8.98E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.607E-05 | 8.565E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 3.121E-05 | 8.974E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 7.179E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 7.87E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.679E-07 | 6.544E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 4.887E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.998E-06 | 4.885E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.768E-06 | 4.134E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 9.163E-06 | 4.477E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 1.119E-05 | 4.555E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.119E-05 | 3.905E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.154E-05 | 3.524E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.206E-05 | 3.273E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.371E-05 | 3.349E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.371E-05 | 3.045E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 3.826E-07 | 9.204E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 6.12E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 4.08E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.648E-06 | 5.804E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.306E-05 | 6.283E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 1.422E-05 | 5.704E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 4.889E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 4.944E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 4.593E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.828E-05 | 4.398E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.828E-05 | 3.998E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.679E-07 | 6.544E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 4.887E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.998E-06 | 4.885E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.768E-06 | 4.134E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 9.163E-06 | 4.477E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 1.119E-05 | 4.555E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.119E-05 | 3.905E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.154E-05 | 3.524E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.206E-05 | 3.273E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.371E-05 | 3.349E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.371E-05 | 3.045E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 3.826E-07 | 9.204E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 6.12E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 4.08E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.648E-06 | 5.804E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.306E-05 | 6.283E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 1.422E-05 | 5.704E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 4.889E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 4.944E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 4.593E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.828E-05 | 4.398E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.828E-05 | 3.998E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 1.28E-07 | 4.571E-04 |
|---|
| protein N-terminus binding | GO:0047485 |  | 2.689E-06 | 4.801E-03 |
|---|
| insulin-like growth factor I binding | GO:0031994 |  | 3.314E-06 | 3.944E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.314E-06 | 2.958E-03 |
|---|
| lamin binding | GO:0005521 |  | 4.969E-06 | 3.549E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 6.954E-06 | 4.139E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.954E-06 | 3.548E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 6.954E-06 | 3.104E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 9.269E-06 | 3.678E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 9.269E-06 | 3.31E-03 |
|---|
| protein phosphorylated amino acid binding | GO:0045309 |  | 9.269E-06 | 3.009E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.044E-06 | 3.809E-03 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.136E-06 | 2.073E-03 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.246E-06 | 1.515E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.646E-06 | 1.501E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.927E-06 | 1.406E-03 |
|---|
| insulin-like growth factor I binding | GO:0031994 |  | 1.316E-05 | 7.999E-03 |
|---|
| lamin binding | GO:0005521 |  | 1.316E-05 | 6.856E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 1.316E-05 | 5.999E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.316E-05 | 5.333E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.972E-05 | 7.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.972E-05 | 6.54E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 3.826E-07 | 9.204E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 6.12E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 4.08E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.648E-06 | 5.804E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.306E-05 | 6.283E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 1.422E-05 | 5.704E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 4.889E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 4.944E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 4.593E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.828E-05 | 4.398E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.828E-05 | 3.998E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 6.041E-07 | 1.389E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.01284463 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.193E-05 | 0.01260993 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 2.343E-05 | 0.0107755 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 8.98E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.607E-05 | 8.565E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 3.121E-05 | 8.974E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 7.179E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 7.87E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 6.041E-07 | 1.389E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.01284463 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.193E-05 | 0.01260993 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 2.343E-05 | 0.0107755 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 8.98E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.607E-05 | 8.565E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 3.121E-05 | 8.974E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 7.179E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 7.87E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 4.833E-07 | 8.907E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.413E-05 | 0.01301779 |
|---|
| regulation of mitosis | GO:0007088 |  | 1.54E-05 | 9.46E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.844E-05 | 0.01310201 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.844E-05 | 0.0104816 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.844E-05 | 8.735E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.844E-05 | 7.487E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 3.696E-05 | 8.515E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 3.978E-05 | 8.146E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.978E-05 | 7.331E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 5.3E-05 | 8.88E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 6.041E-07 | 1.389E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.01284463 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 8.563E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.193E-05 | 0.01260993 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 2.343E-05 | 0.0107755 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 8.98E-03 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.607E-05 | 8.565E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 3.121E-05 | 8.974E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 7.179E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 7.87E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 1.143E-06 | 2.713E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.06E-05 | 0.01257554 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.589E-05 | 0.01256753 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.297E-05 | 0.01362968 |
|---|
| regulation of mitosis | GO:0007088 |  | 2.431E-05 | 0.0115373 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.962E-05 | 0.01171476 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.962E-05 | 0.01004122 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 3.806E-05 | 0.01128918 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.806E-05 | 0.01003483 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.448E-05 | 0.01055602 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 4.754E-05 | 0.01025636 |
|---|



