Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6134-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1546 | 1.085e-02 | 8.215e-03 | 9.301e-02 |
|---|
| IPC-NIBC-129 | 0.2621 | 2.098e-01 | 2.764e-01 | 8.642e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2459 | 1.221e-02 | 1.996e-02 | 3.035e-01 |
|---|
| Loi_GPL570 | 0.2329 | 7.448e-02 | 8.382e-02 | 2.974e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1643 | 1.000e-06 | 1.400e-05 | 2.000e-06 |
|---|
| Parker_GPL1390 | 0.2306 | 5.401e-02 | 6.685e-02 | 4.108e-01 |
|---|
| Parker_GPL887 | 0.0441 | 4.725e-01 | 4.915e-01 | 5.301e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3651 | 8.332e-01 | 7.964e-01 | 9.993e-01 |
|---|
| Schmidt | 0.2376 | 1.737e-02 | 1.990e-02 | 2.124e-01 |
|---|
| Sotiriou | 0.2932 | 1.662e-02 | 1.291e-02 | 2.503e-01 |
|---|
| Van-De-Vijver | 0.1492 | 1.412e-03 | 2.032e-03 | 7.380e-02 |
|---|
| Zhang | 0.2793 | 1.259e-01 | 8.123e-02 | 2.662e-01 |
|---|
| Zhou | 0.4010 | 6.346e-02 | 5.164e-02 | 3.783e-01 |
|---|
Expression data for subnetwork 6134-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
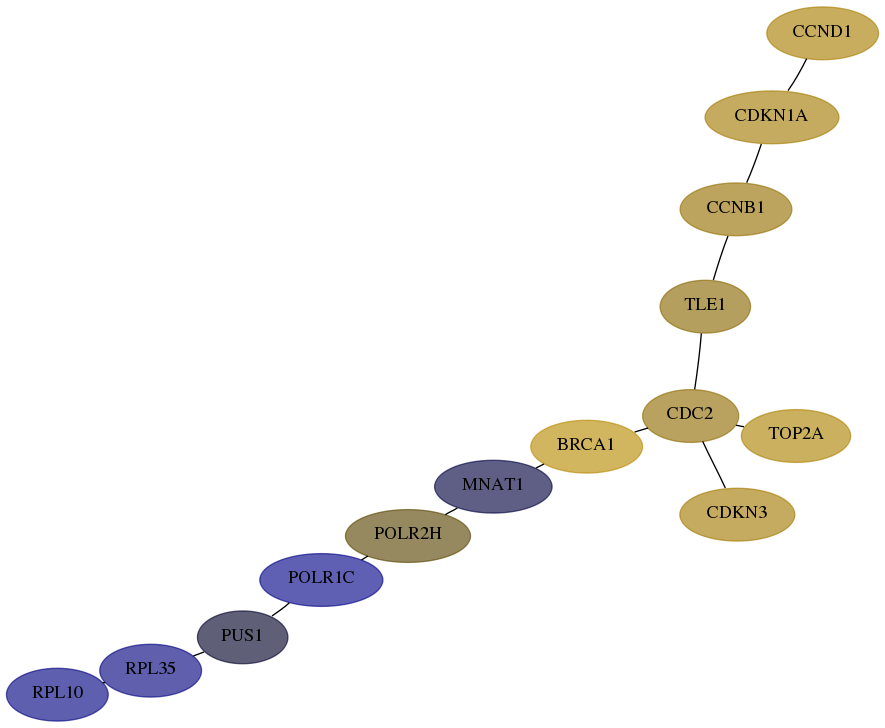
Score for each gene in subnetwork 6134-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| rpl10 |   | 1 | 116 | 294 | 294 | -0.051 | 0.086 | -0.064 | 0.183 | 0.072 | 0.191 | -0.282 | 0.022 | -0.116 | 0.073 | -0.136 | -0.001 | 0.171 |
|---|
| rpl35 |   | 1 | 116 | 294 | 294 | -0.048 | 0.098 | 0.117 | 0.193 | 0.090 | 0.143 | 0.119 | 0.042 | 0.102 | 0.128 | -0.073 | -0.011 | 0.054 |
|---|
| pus1 |   | 1 | 116 | 294 | 294 | -0.006 | 0.172 | 0.189 | -0.093 | 0.107 | 0.216 | 0.074 | 0.110 | 0.165 | 0.118 | undef | 0.050 | 0.195 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| brca1 |   | 2 | 78 | 238 | 230 | 0.192 | 0.052 | 0.112 | 0.022 | 0.125 | -0.124 | 0.305 | 0.113 | 0.077 | 0.168 | 0.031 | 0.064 | 0.209 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| polr1c |   | 1 | 116 | 294 | 294 | -0.059 | 0.185 | 0.148 | 0.301 | 0.075 | 0.129 | 0.076 | 0.150 | 0.283 | 0.069 | -0.015 | 0.068 | 0.284 |
|---|
| polr2h |   | 2 | 78 | 216 | 215 | 0.021 | 0.282 | 0.144 | 0.302 | 0.127 | 0.204 | 0.069 | 0.129 | 0.257 | 0.153 | undef | 0.165 | 0.236 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
| mnat1 |   | 1 | 116 | 294 | 294 | -0.011 | 0.083 | 0.075 | 0.305 | 0.105 | 0.111 | -0.009 | 0.107 | 0.118 | -0.002 | 0.024 | 0.307 | 0.086 |
|---|
GO Enrichment output for subnetwork 6134-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.783E-09 | 2.02E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.259E-07 | 1.448E-04 |
|---|
| interphase | GO:0051325 |  | 1.414E-07 | 1.084E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 2.222E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 1.554E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 7.506E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.958E-05 | 6.434E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.958E-05 | 5.63E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 2.465E-05 | 6.299E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 5.669E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.935E-05 | 6.136E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.726E-09 | 4.217E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.728E-08 | 3.332E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.594E-08 | 6.999E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.504E-06 | 9.186E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.839E-06 | 3.83E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.839E-06 | 3.192E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.166E-06 | 3.199E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.175E-05 | 3.589E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.175E-05 | 3.19E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.175E-05 | 2.871E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.187E-05 | 2.637E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.327E-09 | 5.599E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.14E-08 | 3.777E-05 |
|---|
| interphase | GO:0051325 |  | 3.673E-08 | 2.946E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.1E-07 | 6.615E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.82E-06 | 8.758E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.937E-06 | 3.584E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.937E-06 | 3.072E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 8.937E-06 | 2.688E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.108E-05 | 2.963E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.265E-05 | 3.043E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.34E-05 | 2.93E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.726E-09 | 4.217E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.728E-08 | 3.332E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.594E-08 | 6.999E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.504E-06 | 9.186E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.839E-06 | 3.83E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.839E-06 | 3.192E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.166E-06 | 3.199E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.175E-05 | 3.589E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.175E-05 | 3.19E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.175E-05 | 2.871E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.187E-05 | 2.637E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.327E-09 | 5.599E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.14E-08 | 3.777E-05 |
|---|
| interphase | GO:0051325 |  | 3.673E-08 | 2.946E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.1E-07 | 6.615E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.82E-06 | 8.758E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.937E-06 | 3.584E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.937E-06 | 3.072E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 8.937E-06 | 2.688E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.108E-05 | 2.963E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.265E-05 | 3.043E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.34E-05 | 2.93E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.62E-10 | 1.65E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.864E-09 | 1.047E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.997E-08 | 3.568E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.823E-08 | 5.199E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.818E-08 | 6.298E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.623E-07 | 2.156E-04 |
|---|
| microtubule organizing center part | GO:0044450 |  | 2.378E-06 | 1.213E-03 |
|---|
| DNA-directed RNA polymerase activity | GO:0003899 |  | 3.161E-06 | 1.411E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 3.455E-06 | 1.371E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.488E-06 | 2.317E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.176E-06 | 2.329E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.015E-09 | 1.83E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.333E-08 | 9.727E-05 |
|---|
| interphase | GO:0051325 |  | 6.033E-08 | 7.336E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.032E-07 | 9.416E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.479E-07 | 1.809E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 6.677E-07 | 4.06E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.026E-06 | 5.344E-04 |
|---|
| response to calcium ion | GO:0051592 |  | 8.128E-06 | 3.707E-03 |
|---|
| microtubule organizing center part | GO:0044450 |  | 8.962E-06 | 3.633E-03 |
|---|
| DNA-directed RNA polymerase activity | GO:0003899 |  | 1.517E-05 | 5.532E-03 |
|---|
| cytosolic large ribosomal subunit | GO:0022625 |  | 1.517E-05 | 5.029E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.327E-09 | 5.599E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.14E-08 | 3.777E-05 |
|---|
| interphase | GO:0051325 |  | 3.673E-08 | 2.946E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.1E-07 | 6.615E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.82E-06 | 8.758E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.937E-06 | 3.584E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.937E-06 | 3.072E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 8.937E-06 | 2.688E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.108E-05 | 2.963E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.265E-05 | 3.043E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.34E-05 | 2.93E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.783E-09 | 2.02E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.259E-07 | 1.448E-04 |
|---|
| interphase | GO:0051325 |  | 1.414E-07 | 1.084E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 2.222E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 1.554E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 7.506E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.958E-05 | 6.434E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.958E-05 | 5.63E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 2.465E-05 | 6.299E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 5.669E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.935E-05 | 6.136E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.783E-09 | 2.02E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.259E-07 | 1.448E-04 |
|---|
| interphase | GO:0051325 |  | 1.414E-07 | 1.084E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 2.222E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 1.554E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 7.506E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.958E-05 | 6.434E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.958E-05 | 5.63E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 2.465E-05 | 6.299E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 5.669E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.935E-05 | 6.136E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 8.543E-06 | 0.01574429 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.181E-05 | 0.01088458 |
|---|
| postreplication repair | GO:0006301 |  | 1.181E-05 | 7.256E-03 |
|---|
| regulation of transcription from RNA polymerase III promoter | GO:0006359 |  | 1.181E-05 | 5.442E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.181E-05 | 4.354E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 2.478E-05 | 7.61E-03 |
|---|
| negative regulation of lipid metabolic process | GO:0045833 |  | 2.478E-05 | 6.523E-03 |
|---|
| regulation of fatty acid metabolic process | GO:0019217 |  | 3.302E-05 | 7.606E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.242E-05 | 8.687E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 5.3E-05 | 9.768E-03 |
|---|
| regulation of lipid biosynthetic process | GO:0046890 |  | 5.3E-05 | 8.88E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.783E-09 | 2.02E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.259E-07 | 1.448E-04 |
|---|
| interphase | GO:0051325 |  | 1.414E-07 | 1.084E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 2.222E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 1.554E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 7.506E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.958E-05 | 6.434E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.958E-05 | 5.63E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 2.465E-05 | 6.299E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 5.669E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.935E-05 | 6.136E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.552E-09 | 2.029E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.067E-07 | 1.266E-04 |
|---|
| interphase | GO:0051325 |  | 1.197E-07 | 9.465E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.523E-07 | 2.09E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.92E-06 | 1.386E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.77E-05 | 7.002E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.77E-05 | 6.002E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.77E-05 | 5.252E-03 |
|---|
| regulation of centrosome duplication | GO:0010824 |  | 1.77E-05 | 4.668E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.473E-05 | 5.868E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.653E-05 | 5.724E-03 |
|---|



