Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10456-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2219 | 1.490e-02 | 1.177e-02 | 1.193e-01 |
|---|
| IPC-NIBC-129 | 0.2450 | 6.118e-02 | 8.722e-02 | 5.030e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2305 | 2.399e-02 | 3.694e-02 | 4.269e-01 |
|---|
| Loi_GPL570 | 0.2381 | 6.921e-02 | 7.806e-02 | 2.814e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1603 | 4.000e-06 | 4.100e-05 | 1.400e-05 |
|---|
| Parker_GPL1390 | 0.1992 | 5.342e-02 | 6.616e-02 | 4.084e-01 |
|---|
| Parker_GPL887 | 0.0655 | 3.141e-01 | 3.324e-01 | 3.358e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3692 | 7.177e-01 | 6.629e-01 | 9.959e-01 |
|---|
| Schmidt | 0.2383 | 1.798e-02 | 2.065e-02 | 2.175e-01 |
|---|
| Sotiriou | 0.2881 | 1.810e-02 | 1.409e-02 | 2.622e-01 |
|---|
| Van-De-Vijver | 0.2421 | 9.420e-04 | 1.356e-03 | 5.640e-02 |
|---|
| Zhang | 0.2772 | 1.337e-01 | 8.765e-02 | 2.799e-01 |
|---|
| Zhou | 0.4237 | 6.110e-02 | 4.915e-02 | 3.677e-01 |
|---|
Expression data for subnetwork 10456-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
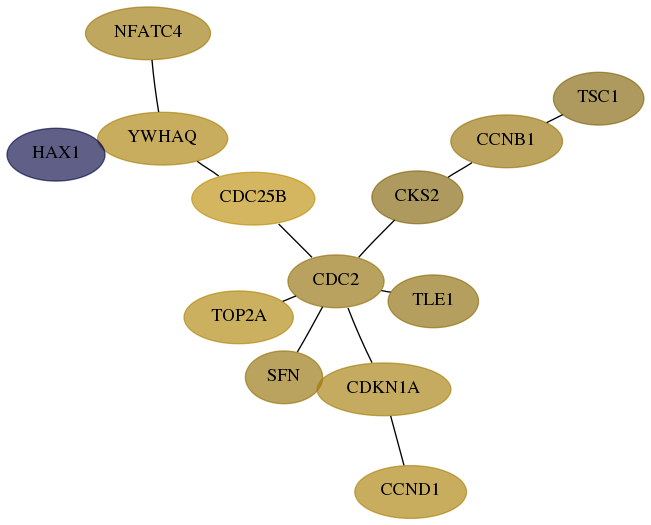
Score for each gene in subnetwork 10456-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| cks2 |   | 12 | 18 | 34 | 32 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.080 | 0.284 | 0.383 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc25b |   | 4 | 50 | 140 | 128 | 0.197 | 0.107 | 0.134 | 0.223 | 0.092 | 0.194 | 0.071 | 0.200 | 0.067 | 0.115 | undef | 0.048 | 0.153 |
|---|
| ywhaq |   | 5 | 40 | 1 | 9 | 0.129 | 0.110 | 0.049 | 0.116 | 0.071 | 0.160 | -0.210 | 0.038 | 0.007 | 0.054 | 0.072 | 0.138 | 0.187 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hax1 |   | 1 | 116 | 240 | 241 | -0.011 | 0.153 | 0.024 | -0.035 | 0.118 | -0.156 | 0.145 | 0.267 | 0.031 | 0.128 | 0.073 | 0.046 | 0.185 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
GO Enrichment output for subnetwork 10456-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 2.607E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 2.861E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 2.352E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 5.782E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.956E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.466E-10 | 2.068E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.952E-07 | 1.216E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 8.609E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 3.649E-03 |
|---|
| female meiosis | GO:0007143 |  | 5.975E-06 | 2.92E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.975E-06 | 2.433E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.076E-06 | 2.12E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 1.984E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.959E-06 | 2.432E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.959E-06 | 2.189E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 2.482E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.929E-09 | 4.64E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 1.964E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.795E-06 | 1.439E-03 |
|---|
| interphase | GO:0051325 |  | 2.032E-06 | 1.223E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 4.004E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 3.337E-03 |
|---|
| female meiosis | GO:0007143 |  | 8.321E-06 | 2.86E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.321E-06 | 2.502E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.321E-06 | 2.224E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.948E-06 | 2.393E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.064E-05 | 2.326E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.466E-10 | 2.068E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.952E-07 | 1.216E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 8.609E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 3.649E-03 |
|---|
| female meiosis | GO:0007143 |  | 5.975E-06 | 2.92E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.975E-06 | 2.433E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.076E-06 | 2.12E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 1.984E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.959E-06 | 2.432E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.959E-06 | 2.189E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 2.482E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.929E-09 | 4.64E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 1.964E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.795E-06 | 1.439E-03 |
|---|
| interphase | GO:0051325 |  | 2.032E-06 | 1.223E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 4.004E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 3.337E-03 |
|---|
| female meiosis | GO:0007143 |  | 8.321E-06 | 2.86E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.321E-06 | 2.502E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.321E-06 | 2.224E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.948E-06 | 2.393E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.064E-05 | 2.326E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.297E-11 | 2.963E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.851E-10 | 5.09E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.02E-09 | 1.214E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.63E-09 | 2.348E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 1.647E-05 |
|---|
| protein N-terminus binding | GO:0047485 |  | 4.814E-08 | 2.865E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.495E-07 | 7.626E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.789E-07 | 1.245E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.264E-07 | 1.692E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.337E-06 | 4.774E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.655E-06 | 5.373E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.394E-10 | 5.085E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.553E-10 | 2.833E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 5.878E-10 | 7.147E-07 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.211E-09 | 1.104E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 1.067E-05 |
|---|
| protein N-terminus binding | GO:0047485 |  | 2.674E-08 | 1.626E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.457E-07 | 7.595E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.457E-07 | 6.646E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.258E-07 | 9.151E-05 |
|---|
| interphase | GO:0051325 |  | 2.492E-07 | 9.089E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.546E-06 | 5.128E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.929E-09 | 4.64E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 1.964E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.795E-06 | 1.439E-03 |
|---|
| interphase | GO:0051325 |  | 2.032E-06 | 1.223E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 4.004E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 3.337E-03 |
|---|
| female meiosis | GO:0007143 |  | 8.321E-06 | 2.86E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.321E-06 | 2.502E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.321E-06 | 2.224E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.948E-06 | 2.393E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.064E-05 | 2.326E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 2.607E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 2.861E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 2.352E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 5.782E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.956E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 2.607E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 2.861E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 2.352E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 5.782E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.956E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.923E-06 | 0.01828869 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.518E-05 | 0.01399039 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 2.137E-05 | 0.01313057 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.276E-05 | 0.01048567 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.276E-05 | 8.389E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.276E-05 | 6.99E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.276E-05 | 5.992E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 3.184E-05 | 7.335E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.184E-05 | 6.52E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 4.242E-05 | 7.819E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.242E-05 | 7.108E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 2.607E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 2.861E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 2.352E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 5.782E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.956E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.593E-09 | 8.527E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.773E-06 | 2.103E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.839E-06 | 2.245E-03 |
|---|
| interphase | GO:0051325 |  | 3.109E-06 | 1.844E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.277E-05 | 6.059E-03 |
|---|
| female meiosis | GO:0007143 |  | 1.277E-05 | 5.049E-03 |
|---|
| oocyte maturation | GO:0001556 |  | 1.277E-05 | 4.328E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.277E-05 | 3.787E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.277E-05 | 3.366E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.506E-05 | 3.574E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.735E-05 | 3.742E-03 |
|---|



