Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7407-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1925 | 1.310e-02 | 1.568e-02 | 1.211e-01 |
|---|
| IPC-NIBC-129 | 0.2673 | 2.686e-02 | 3.271e-02 | 2.876e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2352 | 2.897e-03 | 5.053e-03 | 1.179e-01 |
|---|
| Loi_GPL570 | 0.3006 | 4.851e-02 | 4.815e-02 | 2.492e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1464 | 6.000e-06 | 3.900e-05 | 3.200e-05 |
|---|
| Parker_GPL1390 | 0.3031 | 1.091e-01 | 1.096e-01 | 5.637e-01 |
|---|
| Parker_GPL887 | 0.0779 | 7.314e-01 | 7.550e-01 | 7.873e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3734 | 1.064e-01 | 7.858e-02 | 5.379e-01 |
|---|
| Schmidt | 0.2473 | 6.865e-03 | 9.967e-03 | 9.725e-02 |
|---|
| Sotiriou | 0.3381 | 5.786e-03 | 2.084e-03 | 1.125e-01 |
|---|
| Wang | 0.0515 | 1.376e-01 | 9.985e-02 | 4.477e-01 |
|---|
| Zhang | 0.2651 | 5.997e-01 | 5.545e-01 | 8.778e-01 |
|---|
| Zhou | 0.3622 | 5.397e-02 | 5.638e-02 | 3.395e-01 |
|---|
Expression data for subnetwork 7407-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
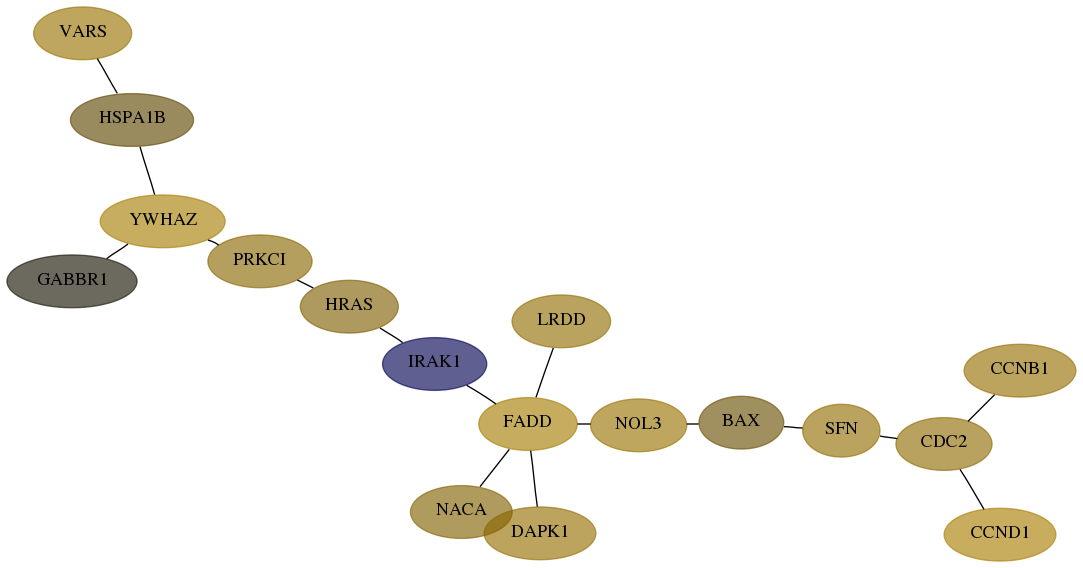
Score for each gene in subnetwork 7407-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| irak1 |   | 3 | 79 | 120 | 112 | -0.016 | 0.242 | 0.007 | 0.133 | 0.108 | 0.303 | 0.257 | 0.133 | 0.177 | 0.084 | -0.046 | -0.026 | 0.081 |
|---|
| vars |   | 1 | 164 | 313 | 319 | 0.088 | 0.254 | 0.146 | -0.113 | 0.062 | 0.236 | 0.104 | 0.249 | 0.110 | 0.093 | 0.038 | -0.087 | 0.077 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| prkci |   | 12 | 20 | 31 | 19 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | -0.054 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hras |   | 7 | 38 | 88 | 75 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.001 | 0.003 | 0.197 |
|---|
| gabbr1 |   | 6 | 43 | 88 | 77 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.011 | 0.096 | 0.116 |
|---|
| lrdd |   | 2 | 104 | 64 | 71 | 0.075 | 0.117 | -0.015 | 0.179 | 0.068 | 0.006 | 0.302 | 0.104 | 0.152 | 0.007 | -0.028 | 0.108 | 0.085 |
|---|
| hspa1b |   | 1 | 164 | 313 | 319 | 0.024 | 0.027 | 0.052 | -0.016 | 0.132 | 0.250 | undef | -0.016 | 0.144 | 0.099 | 0.058 | -0.004 | 0.166 |
|---|
| bax |   | 9 | 27 | 17 | 15 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 | 0.235 |
|---|
| ywhaz |   | 11 | 21 | 31 | 20 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | -0.026 | 0.103 | 0.213 |
|---|
| fadd |   | 5 | 50 | 64 | 58 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.119 | 0.084 | 0.300 | 0.070 | 0.116 | 0.100 | 0.032 | 0.243 |
|---|
| nol3 |   | 3 | 79 | 228 | 221 | 0.089 | 0.110 | 0.102 | -0.119 | 0.094 | 0.021 | 0.040 | 0.125 | -0.023 | 0.016 | 0.180 | 0.059 | 0.069 |
|---|
| dapk1 |   | 3 | 79 | 120 | 112 | 0.082 | 0.188 | -0.078 | -0.063 | 0.067 | 0.083 | -0.195 | 0.267 | 0.117 | 0.059 | -0.061 | 0.142 | 0.230 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| naca |   | 4 | 60 | 120 | 108 | 0.052 | 0.141 | 0.119 | 0.132 | 0.102 | -0.017 | -0.128 | 0.081 | 0.076 | 0.100 | 0.129 | 0.147 | 0.138 |
|---|
GO Enrichment output for subnetwork 7407-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.779E-07 | 2.045E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.452E-07 | 1.88E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 4.287E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.157E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 1.954E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 4.648E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.187E-05 | 5.589E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.633E-05 | 6.055E-03 |
|---|
| amino acid activation | GO:0043038 |  | 3.406E-05 | 7.122E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.126E-08 | 2.75E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.37E-08 | 5.338E-05 |
|---|
| response to protein stimulus | GO:0051789 |  | 6.05E-08 | 4.927E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.2E-07 | 1.955E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 6.252E-07 | 3.055E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.479E-06 | 6.024E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.882E-06 | 1.006E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.882E-06 | 8.799E-04 |
|---|
| sperm motility | GO:0030317 |  | 3.625E-06 | 9.841E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.473E-06 | 1.337E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 7.855E-06 | 1.744E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.312E-08 | 3.158E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.147E-08 | 4.989E-05 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.851E-08 | 4.692E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.73E-07 | 2.244E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.288E-07 | 3.507E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.479E-06 | 5.931E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.618E-06 | 8.997E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.972E-06 | 8.939E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.972E-06 | 7.946E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 6.375E-06 | 1.534E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 9.149E-06 | 2.001E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.126E-08 | 2.75E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.37E-08 | 5.338E-05 |
|---|
| response to protein stimulus | GO:0051789 |  | 6.05E-08 | 4.927E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.2E-07 | 1.955E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 6.252E-07 | 3.055E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.479E-06 | 6.024E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.882E-06 | 1.006E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.882E-06 | 8.799E-04 |
|---|
| sperm motility | GO:0030317 |  | 3.625E-06 | 9.841E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.473E-06 | 1.337E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 7.855E-06 | 1.744E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.312E-08 | 3.158E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.147E-08 | 4.989E-05 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.851E-08 | 4.692E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.73E-07 | 2.244E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.288E-07 | 3.507E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.479E-06 | 5.931E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.618E-06 | 8.997E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.972E-06 | 8.939E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.972E-06 | 7.946E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 6.375E-06 | 1.534E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 9.149E-06 | 2.001E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 5.864E-08 | 2.094E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 6.418E-07 | 1.146E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 9.125E-07 | 1.086E-03 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 3.774E-06 | 3.369E-03 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 9.376E-06 | 6.696E-03 |
|---|
| regulation of neuron apoptosis | GO:0043523 |  | 1.45E-05 | 8.63E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.654E-05 | 8.439E-03 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 1.654E-05 | 7.384E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.654E-05 | 6.564E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.654E-05 | 5.907E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.996E-05 | 6.48E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 2.861E-08 | 1.044E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 2.874E-08 | 5.243E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.854E-07 | 2.254E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.749E-07 | 6.156E-04 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 1.847E-06 | 1.347E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.354E-06 | 3.255E-03 |
|---|
| regulation of neuron apoptosis | GO:0043523 |  | 6.641E-06 | 3.461E-03 |
|---|
| lymphocyte apoptosis | GO:0070227 |  | 6.839E-06 | 3.118E-03 |
|---|
| NF-kappaB-inducing kinase activity | GO:0004704 |  | 6.839E-06 | 2.772E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 6.839E-06 | 2.495E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.025E-05 | 3.4E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.312E-08 | 3.158E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.147E-08 | 4.989E-05 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.851E-08 | 4.692E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.73E-07 | 2.244E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.288E-07 | 3.507E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.479E-06 | 5.931E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.618E-06 | 8.997E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.972E-06 | 8.939E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.972E-06 | 7.946E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 6.375E-06 | 1.534E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 9.149E-06 | 2.001E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.779E-07 | 2.045E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.452E-07 | 1.88E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 4.287E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.157E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 1.954E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 4.648E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.187E-05 | 5.589E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.633E-05 | 6.055E-03 |
|---|
| amino acid activation | GO:0043038 |  | 3.406E-05 | 7.122E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.779E-07 | 2.045E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.452E-07 | 1.88E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 4.287E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.157E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 1.954E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 4.648E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.187E-05 | 5.589E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.633E-05 | 6.055E-03 |
|---|
| amino acid activation | GO:0043038 |  | 3.406E-05 | 7.122E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.779E-07 | 2.045E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.452E-07 | 1.88E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 4.287E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.157E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 1.954E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 4.648E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.187E-05 | 5.589E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.633E-05 | 6.055E-03 |
|---|
| amino acid activation | GO:0043038 |  | 3.406E-05 | 7.122E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.779E-07 | 2.045E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.452E-07 | 1.88E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 4.287E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.157E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 1.954E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 4.648E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.187E-05 | 5.589E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.633E-05 | 6.055E-03 |
|---|
| amino acid activation | GO:0043038 |  | 3.406E-05 | 7.122E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.554E-08 | 1.081E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.356E-07 | 2.796E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 3.195E-07 | 2.527E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 9.938E-07 | 5.896E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.054E-06 | 1.449E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.876E-06 | 2.719E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.155E-05 | 3.917E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.986E-05 | 5.891E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.634E-05 | 6.946E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 2.828E-05 | 6.71E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 3.409E-05 | 7.354E-03 |
|---|



