Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5155-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2354 | 9.227e-03 | 1.102e-02 | 9.327e-02 |
|---|
| IPC-NIBC-129 | 0.2526 | 3.656e-02 | 4.557e-02 | 3.638e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2522 | 7.410e-03 | 1.175e-02 | 2.192e-01 |
|---|
| Loi_GPL570 | 0.2570 | 5.931e-02 | 5.890e-02 | 2.887e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1528 | 3.000e-06 | 2.100e-05 | 1.100e-05 |
|---|
| Parker_GPL1390 | 0.2809 | 5.973e-02 | 5.679e-02 | 4.155e-01 |
|---|
| Parker_GPL887 | 0.2310 | 2.919e-01 | 3.255e-01 | 2.945e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2808 | 9.268e-02 | 6.696e-02 | 4.948e-01 |
|---|
| Schmidt | 0.2569 | 6.725e-03 | 9.772e-03 | 9.565e-02 |
|---|
| Sotiriou | 0.3393 | 3.720e-03 | 1.195e-03 | 8.279e-02 |
|---|
| Wang | 0.1012 | 1.243e-01 | 8.866e-02 | 4.202e-01 |
|---|
| Zhang | 0.2786 | 3.147e-01 | 2.578e-01 | 6.029e-01 |
|---|
| Zhou | 0.3889 | 1.289e-01 | 1.450e-01 | 6.486e-01 |
|---|
Expression data for subnetwork 5155-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
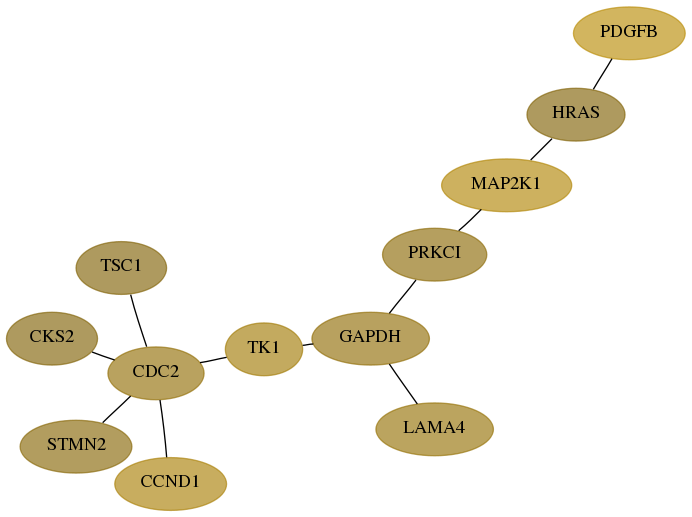
Score for each gene in subnetwork 5155-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| pdgfb |   | 2 | 104 | 161 | 156 | 0.186 | -0.069 | 0.089 | 0.269 | 0.087 | 0.107 | -0.031 | 0.163 | 0.038 | 0.139 | -0.163 | 0.037 | 0.170 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| prkci |   | 12 | 20 | 31 | 19 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | -0.054 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| lama4 |   | 5 | 50 | 136 | 121 | 0.079 | 0.190 | 0.023 | -0.039 | 0.041 | 0.126 | -0.167 | -0.168 | -0.007 | -0.089 | 0.200 | 0.057 | 0.089 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| gapdh |   | 6 | 43 | 136 | 117 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.172 | -0.253 | 0.288 | 0.064 | 0.213 | 0.056 | -0.088 | 0.235 |
|---|
| tk1 |   | 6 | 43 | 1 | 7 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.105 | -0.100 | 0.257 | 0.204 | 0.142 | 0.064 | 0.164 | 0.222 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hras |   | 7 | 38 | 88 | 75 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.001 | 0.003 | 0.197 |
|---|
| map2k1 |   | 2 | 104 | 120 | 118 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | -0.043 | undef | 0.230 |
|---|
GO Enrichment output for subnetwork 5155-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 0.03799068 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 0.01899534 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.652E-05 | 0.01266356 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.476E-05 | 0.0142351 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01137896 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.463E-05 | 9.957E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 3.463E-05 | 7.241E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 0.01459787 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.975E-06 | 7.299E-03 |
|---|
| glycolysis | GO:0006096 |  | 6.496E-06 | 5.29E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 8.959E-06 | 5.472E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 8.959E-06 | 4.377E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.959E-06 | 3.648E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.18E-05 | 4.118E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.245E-05 | 3.802E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 4.535E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.671E-05 | 4.081E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.671E-05 | 3.71E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 0.01589185 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 7.946E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.605E-06 | 5.297E-03 |
|---|
| glycolysis | GO:0006096 |  | 7.006E-06 | 4.214E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 9.903E-06 | 4.765E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 9.903E-06 | 3.971E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.288E-05 | 4.428E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.435E-05 | 4.316E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 4.937E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.847E-05 | 4.443E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.847E-05 | 4.039E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 0.01459787 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.975E-06 | 7.299E-03 |
|---|
| glycolysis | GO:0006096 |  | 6.496E-06 | 5.29E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 8.959E-06 | 5.472E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 8.959E-06 | 4.377E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.959E-06 | 3.648E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.18E-05 | 4.118E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.245E-05 | 3.802E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 4.535E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.671E-05 | 4.081E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.671E-05 | 3.71E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 0.01589185 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 7.946E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.605E-06 | 5.297E-03 |
|---|
| glycolysis | GO:0006096 |  | 7.006E-06 | 4.214E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 9.903E-06 | 4.765E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 9.903E-06 | 3.971E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.288E-05 | 4.428E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.435E-05 | 4.316E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 4.937E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.847E-05 | 4.443E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.847E-05 | 4.039E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.091E-08 | 1.818E-04 |
|---|
| site of polarized growth | GO:0030427 |  | 4.731E-07 | 8.447E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.061E-06 | 6.024E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 4.518E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.952E-06 | 4.251E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 4.517E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 7.589E-06 | 3.871E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 7.589E-06 | 3.387E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.062E-05 | 4.214E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.062E-05 | 3.792E-03 |
|---|
| laminin-1 complex | GO:0005606 |  | 1.062E-05 | 3.448E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.481E-08 | 3.459E-04 |
|---|
| site of polarized growth | GO:0030427 |  | 5.964E-07 | 1.088E-03 |
|---|
| platelet-derived growth factor binding | GO:0048407 |  | 4.447E-06 | 5.407E-03 |
|---|
| platelet dense granule | GO:0042827 |  | 4.447E-06 | 4.055E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 4.447E-06 | 3.244E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 2.704E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 2.317E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.344E-06 | 2.437E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.667E-06 | 2.702E-03 |
|---|
| regulation of platelet activation | GO:0010543 |  | 6.667E-06 | 2.432E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.667E-06 | 2.211E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 0.01589185 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 7.946E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.605E-06 | 5.297E-03 |
|---|
| glycolysis | GO:0006096 |  | 7.006E-06 | 4.214E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 9.903E-06 | 4.765E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 9.903E-06 | 3.971E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.288E-05 | 4.428E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.435E-05 | 4.316E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 4.937E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.847E-05 | 4.443E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.847E-05 | 4.039E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 0.03799068 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 0.01899534 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.652E-05 | 0.01266356 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.476E-05 | 0.0142351 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01137896 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.463E-05 | 9.957E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 3.463E-05 | 7.241E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 0.03799068 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 0.01899534 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.652E-05 | 0.01266356 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.476E-05 | 0.0142351 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01137896 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.463E-05 | 9.957E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 3.463E-05 | 7.241E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 0.03799068 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 0.01899534 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.652E-05 | 0.01266356 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.476E-05 | 0.0142351 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01137896 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.463E-05 | 9.957E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 3.463E-05 | 7.241E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 0.03799068 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 0.01899534 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.652E-05 | 0.01266356 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.476E-05 | 0.0142351 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01137896 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.463E-05 | 9.957E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 3.463E-05 | 7.241E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 2.227E-08 | 5.285E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 7.782E-08 | 9.233E-05 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 1.864E-07 | 1.475E-04 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 1.864E-07 | 1.106E-04 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 2.661E-07 | 1.263E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.661E-07 | 1.052E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 4.869E-07 | 1.651E-04 |
|---|
| glycolysis | GO:0006096 |  | 5.473E-07 | 1.623E-04 |
|---|
| negative regulation of lipid metabolic process | GO:0045833 |  | 6.324E-07 | 1.667E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 6.324E-07 | 1.501E-04 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.235E-06 | 2.664E-04 |
|---|



