Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 23438-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2003 | 7.730e-03 | 9.223e-03 | 8.158e-02 |
|---|
| IPC-NIBC-129 | 0.2923 | 4.407e-02 | 5.566e-02 | 4.154e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2608 | 8.177e-03 | 1.284e-02 | 2.330e-01 |
|---|
| Loi_GPL570 | 0.2524 | 3.433e-02 | 3.404e-02 | 1.918e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1567 | 1.000e-06 | 9.000e-06 | 2.000e-06 |
|---|
| Parker_GPL1390 | 0.2674 | 9.785e-02 | 9.735e-02 | 5.352e-01 |
|---|
| Parker_GPL887 | 0.0692 | 7.651e-01 | 7.866e-01 | 8.208e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3485 | 2.365e-01 | 1.981e-01 | 7.972e-01 |
|---|
| Schmidt | 0.2697 | 6.914e-03 | 1.004e-02 | 9.780e-02 |
|---|
| Sotiriou | 0.3376 | 3.103e-03 | 9.500e-04 | 7.279e-02 |
|---|
| Wang | 0.2028 | 1.167e-01 | 8.235e-02 | 4.037e-01 |
|---|
| Zhang | 0.1853 | 7.535e-02 | 4.667e-02 | 1.863e-01 |
|---|
| Zhou | 0.3999 | 6.840e-02 | 7.308e-02 | 4.150e-01 |
|---|
Expression data for subnetwork 23438-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
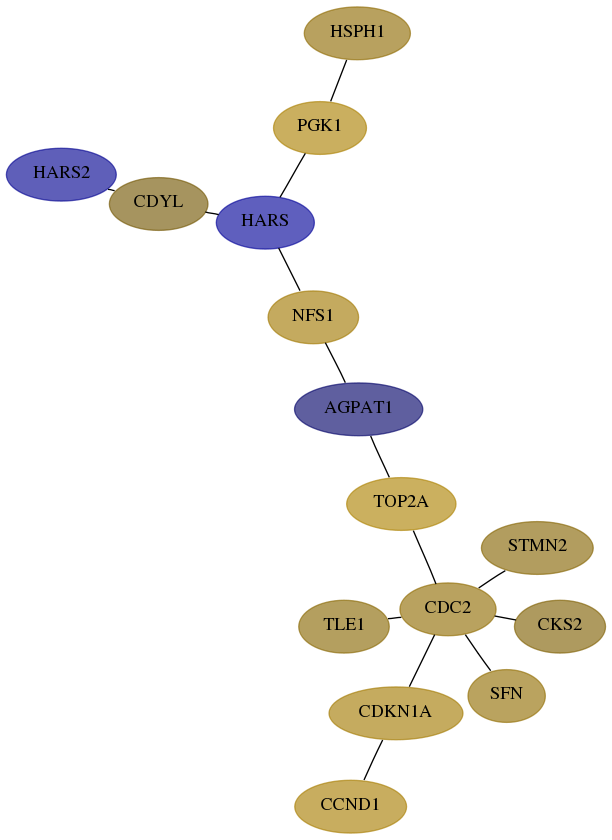
Score for each gene in subnetwork 23438-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| hars2 |   | 1 | 164 | 192 | 200 | -0.075 | 0.082 | 0.119 | 0.195 | 0.126 | 0.017 | -0.087 | 0.089 | 0.005 | 0.105 | -0.020 | -0.122 | 0.370 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| pgk1 |   | 5 | 50 | 53 | 47 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | 0.110 | 0.054 | 0.141 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| cdyl |   | 1 | 164 | 192 | 200 | 0.036 | 0.180 | 0.113 | 0.200 | 0.083 | 0.164 | 0.242 | 0.042 | -0.024 | 0.178 | 0.070 | -0.131 | 0.229 |
|---|
| agpat1 |   | 3 | 79 | 192 | 178 | -0.027 | 0.104 | 0.123 | 0.096 | 0.093 | 0.167 | 0.097 | 0.028 | 0.097 | 0.101 | -0.079 | 0.018 | -0.007 |
|---|
| hsph1 |   | 4 | 60 | 53 | 50 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | 0.169 | 0.094 | 0.357 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| top2a |   | 11 | 21 | 93 | 76 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 | 0.273 |
|---|
| hars |   | 1 | 164 | 192 | 200 | -0.088 | 0.134 | 0.097 | 0.247 | 0.103 | 0.193 | -0.007 | 0.099 | -0.083 | 0.162 | 0.001 | -0.122 | 0.251 |
|---|
| nfs1 |   | 1 | 164 | 192 | 200 | 0.110 | -0.062 | 0.040 | 0.192 | 0.076 | -0.265 | 0.135 | 0.152 | 0.120 | 0.122 | 0.201 | -0.039 | 0.178 |
|---|
GO Enrichment output for subnetwork 23438-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.672E-08 | 4.222E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.071E-07 | 2.354E-04 |
|---|
| glycolysis | GO:0006096 |  | 6.837E-07 | 3.931E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.575E-06 | 7.247E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.13E-06 | 1.2E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.75E-06 | 1.561E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.657E-06 | 2.201E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.334E-06 | 2.385E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.393E-05 | 3.205E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.13E-05 | 4.453E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.032E-09 | 4.964E-06 |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 7.281E-09 | 8.894E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 7.281E-09 | 5.929E-06 |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 1.455E-08 | 8.889E-06 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.545E-08 | 1.244E-05 |
|---|
| glycolysis | GO:0006096 |  | 9.771E-08 | 3.978E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.197E-07 | 4.177E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.185E-07 | 6.672E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.807E-07 | 1.305E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 7.94E-07 | 1.94E-04 |
|---|
| sulfur amino acid metabolic process | GO:0000096 |  | 1.275E-06 | 2.831E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.929E-09 | 9.454E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.088E-08 | 1.309E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.516E-07 | 1.216E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.787E-07 | 1.075E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.445E-07 | 1.658E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 7.674E-07 | 3.077E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.276E-06 | 4.386E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.096E-06 | 6.304E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 3.132E-06 | 8.373E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.968E-06 | 1.677E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.092E-05 | 2.387E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.032E-09 | 4.964E-06 |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 7.281E-09 | 8.894E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 7.281E-09 | 5.929E-06 |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 1.455E-08 | 8.889E-06 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.545E-08 | 1.244E-05 |
|---|
| glycolysis | GO:0006096 |  | 9.771E-08 | 3.978E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.197E-07 | 4.177E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.185E-07 | 6.672E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.807E-07 | 1.305E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 7.94E-07 | 1.94E-04 |
|---|
| sulfur amino acid metabolic process | GO:0000096 |  | 1.275E-06 | 2.831E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.929E-09 | 9.454E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.088E-08 | 1.309E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.516E-07 | 1.216E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.787E-07 | 1.075E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.445E-07 | 1.658E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 7.674E-07 | 3.077E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.276E-06 | 4.386E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.096E-06 | 6.304E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 3.132E-06 | 8.373E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.968E-06 | 1.677E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.092E-05 | 2.387E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.676E-10 | 5.987E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.169E-10 | 1.28E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 2.561E-09 | 3.049E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 6.594E-09 | 5.886E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.814E-08 | 2.724E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 2.471E-07 | 1.47E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 2.206E-06 | 1.125E-03 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 2.731E-06 | 1.219E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 8.241E-06 | 3.27E-03 |
|---|
| phospholipid:diacylglycerol acyltransferase activity | GO:0046027 |  | 8.371E-06 | 2.989E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.371E-06 | 2.717E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.543E-10 | 1.293E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.239E-10 | 9.556E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.979E-09 | 2.406E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 4.07E-09 | 3.712E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.861E-08 | 2.088E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 2.849E-07 | 1.732E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 3.016E-06 | 1.572E-03 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 3.297E-06 | 1.504E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.354E-06 | 2.17E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.191E-06 | 2.259E-03 |
|---|
| amino acid activation | GO:0043038 |  | 6.641E-06 | 2.202E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.929E-09 | 9.454E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.088E-08 | 1.309E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.516E-07 | 1.216E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.787E-07 | 1.075E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.445E-07 | 1.658E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 7.674E-07 | 3.077E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.276E-06 | 4.386E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.096E-06 | 6.304E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 3.132E-06 | 8.373E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.968E-06 | 1.677E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.092E-05 | 2.387E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.672E-08 | 4.222E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.071E-07 | 2.354E-04 |
|---|
| glycolysis | GO:0006096 |  | 6.837E-07 | 3.931E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.575E-06 | 7.247E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.13E-06 | 1.2E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.75E-06 | 1.561E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.657E-06 | 2.201E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.334E-06 | 2.385E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.393E-05 | 3.205E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.13E-05 | 4.453E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.672E-08 | 4.222E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.071E-07 | 2.354E-04 |
|---|
| glycolysis | GO:0006096 |  | 6.837E-07 | 3.931E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.575E-06 | 7.247E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.13E-06 | 1.2E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.75E-06 | 1.561E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.657E-06 | 2.201E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.334E-06 | 2.385E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.393E-05 | 3.205E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.13E-05 | 4.453E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.672E-08 | 4.222E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.071E-07 | 2.354E-04 |
|---|
| glycolysis | GO:0006096 |  | 6.837E-07 | 3.931E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.575E-06 | 7.247E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.13E-06 | 1.2E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.75E-06 | 1.561E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.657E-06 | 2.201E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.334E-06 | 2.385E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.393E-05 | 3.205E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.13E-05 | 4.453E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.672E-08 | 4.222E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.071E-07 | 2.354E-04 |
|---|
| glycolysis | GO:0006096 |  | 6.837E-07 | 3.931E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.575E-06 | 7.247E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.13E-06 | 1.2E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.75E-06 | 1.561E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.657E-06 | 2.201E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.334E-06 | 2.385E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.393E-05 | 3.205E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.13E-05 | 4.453E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.112E-08 | 5.012E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.776E-08 | 4.48E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 4.506E-07 | 3.564E-04 |
|---|
| glycolysis | GO:0006096 |  | 1.111E-06 | 6.593E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.526E-06 | 1.199E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.697E-06 | 1.858E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.878E-06 | 2.332E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.068E-05 | 3.169E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.206E-05 | 3.18E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.942E-05 | 4.608E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 2.501E-05 | 5.394E-03 |
|---|



