Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 93
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5303 | 0.000e+00 | 0.000e+00 | 2.160e-04 | 0.000e+00 |
|---|
| IPC | 0.4702 | 9.496e-02 | 7.563e-02 | 9.563e-01 | 6.868e-03 |
|---|
| Loi | 0.5143 | 0.000e+00 | 0.000e+00 | 2.272e-03 | 0.000e+00 |
|---|
| Schmidt | 0.2868 | 1.724e-02 | 5.138e-03 | 3.294e-01 | 2.918e-05 |
|---|
| Wang | 0.3376 | 1.360e-04 | 3.000e-06 | 5.223e-02 | 2.131e-11 |
|---|
Expression data for subnetwork 93 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
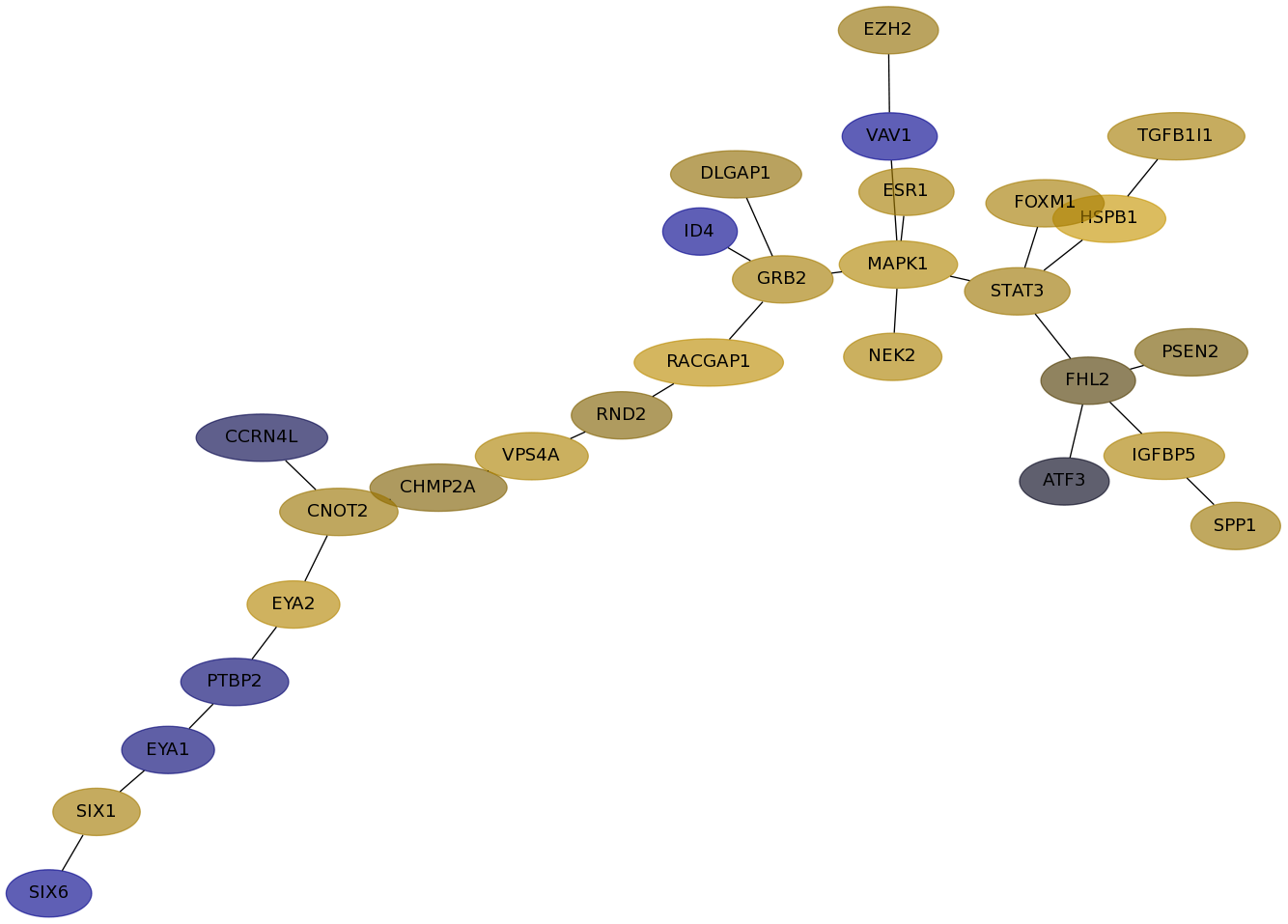
Score for each gene in subnetwork 93 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| DLGAP1 |   | 1 | 59 | 110 | 115 | 0.072 | 0.143 | 0.052 | -0.088 | 0.111 |
|---|
| CHMP2A |   | 1 | 59 | 110 | 115 | 0.050 | 0.019 | 0.209 | 0.071 | -0.011 |
|---|
| PTBP2 |   | 1 | 59 | 110 | 115 | -0.032 | 0.043 | 0.072 | 0.047 | 0.065 |
|---|
| SIX6 |   | 1 | 59 | 110 | 115 | -0.064 | 0.144 | -0.082 | 0.000 | 0.086 |
|---|
| FOXM1 |   | 2 | 27 | 110 | 113 | 0.119 | 0.137 | 0.170 | 0.219 | 0.183 |
|---|
| EYA1 |   | 1 | 59 | 110 | 115 | -0.037 | 0.112 | -0.006 | 0.138 | 0.076 |
|---|
| FHL2 |   | 1 | 59 | 110 | 115 | 0.015 | 0.023 | 0.055 | -0.067 | -0.040 |
|---|
| ATF3 |   | 4 | 6 | 80 | 82 | -0.003 | -0.016 | 0.092 | -0.045 | -0.079 |
|---|
| ID4 |   | 1 | 59 | 110 | 115 | -0.066 | -0.138 | -0.124 | -0.170 | -0.071 |
|---|
| RACGAP1 |   | 3 | 13 | 110 | 111 | 0.199 | 0.154 | 0.232 | 0.232 | 0.337 |
|---|
| EZH2 |   | 1 | 59 | 110 | 115 | 0.076 | 0.106 | 0.107 | 0.222 | 0.277 |
|---|
| GRB2 |   | 7 | 1 | 80 | 80 | 0.121 | 0.037 | 0.087 | 0.182 | -0.037 |
|---|
| CNOT2 |   | 1 | 59 | 110 | 115 | 0.093 | 0.062 | 0.100 | 0.021 | 0.192 |
|---|
| VAV1 |   | 1 | 59 | 110 | 115 | -0.070 | -0.014 | -0.041 | -0.080 | 0.015 |
|---|
| RND2 |   | 1 | 59 | 110 | 115 | 0.051 | 0.042 | 0.081 | 0.097 | 0.188 |
|---|
| SPP1 |   | 3 | 13 | 36 | 37 | 0.096 | 0.208 | 0.125 | -0.007 | 0.149 |
|---|
| SIX1 |   | 1 | 59 | 110 | 115 | 0.113 | 0.156 | 0.121 | 0.168 | 0.089 |
|---|
| IGFBP5 |   | 4 | 6 | 1 | 3 | 0.142 | 0.011 | 0.158 | 0.087 | -0.000 |
|---|
| NEK2 |   | 3 | 13 | 110 | 111 | 0.147 | 0.147 | 0.232 | 0.245 | 0.336 |
|---|
| PSEN2 |   | 1 | 59 | 110 | 115 | 0.042 | 0.054 | 0.186 | 0.218 | 0.118 |
|---|
| VPS4A |   | 1 | 59 | 110 | 115 | 0.147 | 0.092 | 0.081 | 0.173 | 0.104 |
|---|
| HSPB1 |   | 6 | 2 | 1 | 1 | 0.262 | 0.053 | 0.231 | 0.043 | 0.144 |
|---|
| CCRN4L |   | 1 | 59 | 110 | 115 | -0.013 | 0.034 | 0.072 | 0.143 | -0.040 |
|---|
| MAPK1 |   | 5 | 4 | 110 | 110 | 0.157 | -0.023 | -0.125 | -0.115 | 0.092 |
|---|
| ESR1 |   | 6 | 2 | 1 | 1 | 0.125 | 0.032 | 0.114 | -0.064 | 0.048 |
|---|
| STAT3 |   | 2 | 27 | 110 | 113 | 0.100 | -0.078 | -0.046 | 0.077 | -0.189 |
|---|
| EYA2 |   | 1 | 59 | 110 | 115 | 0.164 | 0.054 | 0.119 | 0.097 | 0.174 |
|---|
| TGFB1I1 |   | 1 | 59 | 110 | 115 | 0.119 | 0.041 | -0.103 | -0.159 | 0.146 |
|---|
GO Enrichment output for subnetwork 93 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.193E-07 | 5.043E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 6.371E-07 | 7.326E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 7.643E-07 | 5.86E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 9.668E-07 | 5.559E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 7.837E-06 | 3.605E-03 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 1.743E-05 | 6.681E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 3.263E-05 | 0.01072049 |
|---|
| regulation of neuron differentiation | GO:0045664 |  | 3.974E-05 | 0.01142442 |
|---|
| inner ear morphogenesis | GO:0042472 |  | 6.135E-05 | 0.01567833 |
|---|
| negative regulation of gliogenesis | GO:0014014 |  | 7.973E-05 | 0.0183379 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 8.464E-05 | 0.01769806 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 9.744E-08 | 2.381E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 3.37E-07 | 4.117E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 4.869E-07 | 3.965E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 1.593E-06 | 9.728E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 3.497E-06 | 1.709E-03 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 1.085E-05 | 4.418E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 1.461E-05 | 5.1E-03 |
|---|
| regulation of neuron differentiation | GO:0045664 |  | 3.608E-05 | 0.01101721 |
|---|
| Notch receptor processing | GO:0007220 |  | 4.613E-05 | 0.01252077 |
|---|
| muscle cell migration | GO:0014812 |  | 4.613E-05 | 0.01126869 |
|---|
| metanephros development | GO:0001656 |  | 4.634E-05 | 0.01029096 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 9.124E-08 | 2.195E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.362E-07 | 2.841E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 3.474E-07 | 2.786E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 1.492E-06 | 8.973E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 3.276E-06 | 1.576E-03 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 8.655E-06 | 3.471E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 1.369E-05 | 4.707E-03 |
|---|
| regulation of neuron differentiation | GO:0045664 |  | 2.728E-05 | 8.206E-03 |
|---|
| inner ear morphogenesis | GO:0042472 |  | 3.943E-05 | 0.01053983 |
|---|
| metanephros development | GO:0001656 |  | 4.344E-05 | 0.01045128 |
|---|
| Notch receptor processing | GO:0007220 |  | 4.427E-05 | 9.683E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.193E-07 | 5.043E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 6.371E-07 | 7.326E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 7.643E-07 | 5.86E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 9.668E-07 | 5.559E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 7.837E-06 | 3.605E-03 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 1.743E-05 | 6.681E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 3.263E-05 | 0.01072049 |
|---|
| regulation of neuron differentiation | GO:0045664 |  | 3.974E-05 | 0.01142442 |
|---|
| inner ear morphogenesis | GO:0042472 |  | 6.135E-05 | 0.01567833 |
|---|
| negative regulation of gliogenesis | GO:0014014 |  | 7.973E-05 | 0.0183379 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 8.464E-05 | 0.01769806 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.193E-07 | 5.043E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 6.371E-07 | 7.326E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 7.643E-07 | 5.86E-04 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 9.668E-07 | 5.559E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 7.837E-06 | 3.605E-03 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 1.743E-05 | 6.681E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 3.263E-05 | 0.01072049 |
|---|
| regulation of neuron differentiation | GO:0045664 |  | 3.974E-05 | 0.01142442 |
|---|
| inner ear morphogenesis | GO:0042472 |  | 6.135E-05 | 0.01567833 |
|---|
| negative regulation of gliogenesis | GO:0014014 |  | 7.973E-05 | 0.0183379 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 8.464E-05 | 0.01769806 |
|---|


