Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 63
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5242 | 0.000e+00 | 0.000e+00 | 3.260e-04 | 0.000e+00 |
|---|
| IPC | 0.3833 | 2.159e-01 | 2.082e-01 | 9.659e-01 | 4.341e-02 |
|---|
| Loi | 0.5146 | 0.000e+00 | 0.000e+00 | 2.258e-03 | 0.000e+00 |
|---|
| Schmidt | 0.3123 | 7.153e-03 | 1.554e-03 | 1.930e-01 | 2.146e-06 |
|---|
| Wang | 0.3075 | 7.940e-04 | 3.800e-05 | 1.461e-01 | 4.409e-09 |
|---|
Expression data for subnetwork 63 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
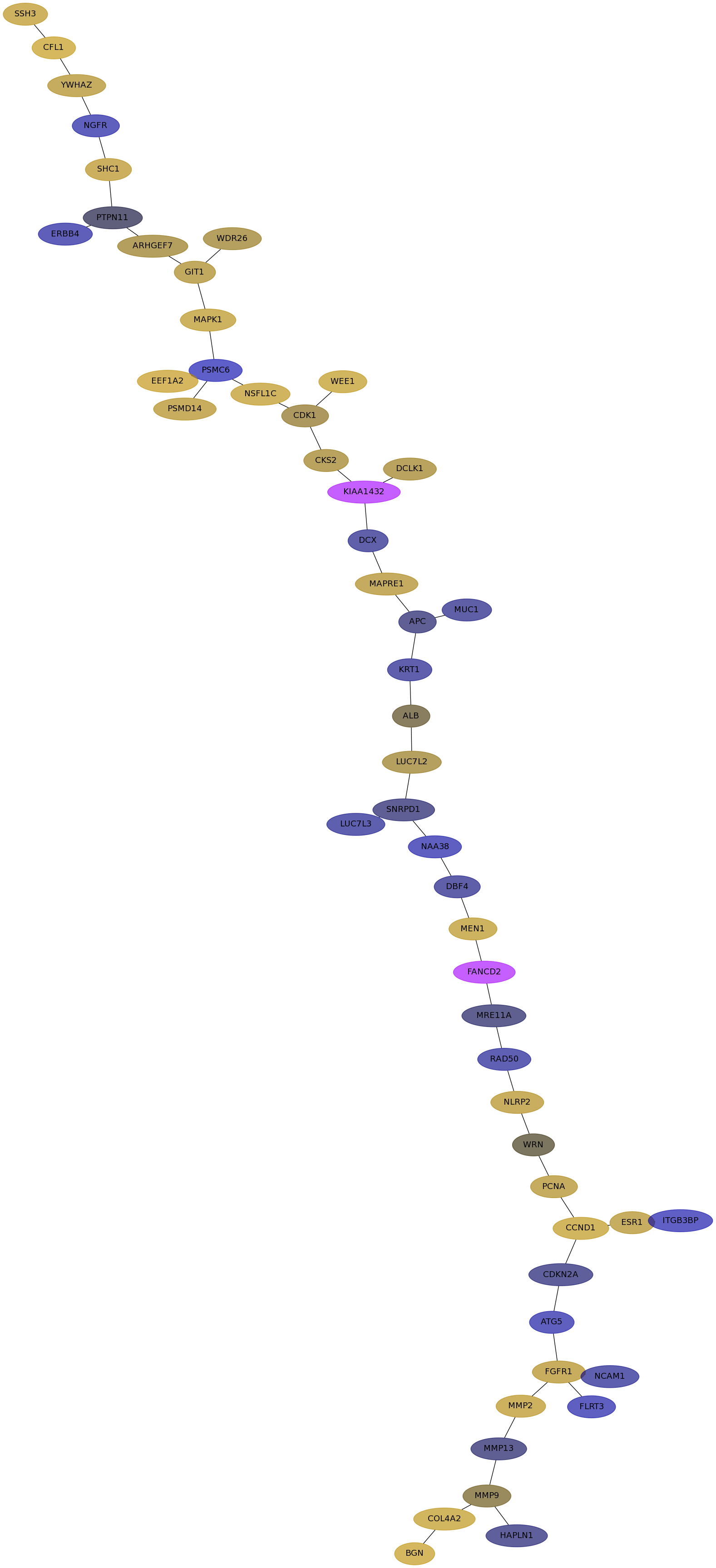
Score for each gene in subnetwork 63 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MMP9 |   | 1 | 59 | 202 | 202 | 0.023 | 0.131 | 0.054 | -0.014 | 0.086 |
|---|
| MAPRE1 |   | 1 | 59 | 202 | 202 | 0.113 | 0.129 | 0.108 | 0.139 | 0.094 |
|---|
| COL4A2 |   | 1 | 59 | 202 | 202 | 0.190 | 0.066 | -0.056 | -0.089 | 0.042 |
|---|
| ARHGEF7 |   | 1 | 59 | 202 | 202 | 0.062 | -0.103 | -0.121 | 0.041 | 0.078 |
|---|
| FANCD2 |   | 1 | 59 | 202 | 202 | undef | 0.135 | 0.059 | undef | undef |
|---|
| DBF4 |   | 1 | 59 | 202 | 202 | -0.040 | 0.098 | 0.145 | 0.084 | 0.198 |
|---|
| WDR26 |   | 1 | 59 | 202 | 202 | 0.066 | 0.119 | 0.173 | 0.193 | 0.171 |
|---|
| APC |   | 1 | 59 | 202 | 202 | -0.020 | 0.141 | 0.062 | 0.033 | 0.067 |
|---|
| GIT1 |   | 1 | 59 | 202 | 202 | 0.102 | -0.032 | 0.059 | 0.112 | -0.070 |
|---|
| DCX |   | 1 | 59 | 202 | 202 | -0.042 | 0.077 | 0.034 | -0.067 | -0.094 |
|---|
| KIAA1432 |   | 1 | 59 | 202 | 202 | undef | 0.132 | 0.034 | undef | undef |
|---|
| ERBB4 |   | 2 | 27 | 36 | 41 | -0.075 | 0.029 | -0.029 | 0.104 | 0.004 |
|---|
| HAPLN1 |   | 2 | 27 | 36 | 41 | -0.025 | 0.142 | 0.086 | -0.085 | 0.083 |
|---|
| CCND1 |   | 4 | 6 | 36 | 36 | 0.189 | 0.059 | 0.145 | 0.009 | 0.146 |
|---|
| WEE1 |   | 2 | 27 | 1 | 10 | 0.186 | -0.013 | -0.003 | 0.043 | 0.146 |
|---|
| PCNA |   | 3 | 13 | 1 | 5 | 0.120 | 0.192 | 0.141 | 0.138 | 0.184 |
|---|
| SNRPD1 |   | 1 | 59 | 202 | 202 | -0.019 | 0.127 | 0.147 | 0.114 | 0.114 |
|---|
| SHC1 |   | 4 | 6 | 167 | 167 | 0.149 | -0.137 | 0.092 | -0.200 | 0.113 |
|---|
| FLRT3 |   | 1 | 59 | 202 | 202 | -0.098 | 0.038 | -0.028 | 0.008 | 0.218 |
|---|
| MMP2 |   | 1 | 59 | 202 | 202 | 0.153 | -0.127 | -0.074 | -0.137 | -0.003 |
|---|
| EEF1A2 |   | 2 | 27 | 36 | 41 | 0.214 | -0.012 | 0.169 | 0.135 | 0.265 |
|---|
| PTPN11 |   | 1 | 59 | 202 | 202 | -0.006 | 0.053 | 0.092 | -0.130 | 0.174 |
|---|
| NSFL1C |   | 1 | 59 | 202 | 202 | 0.177 | 0.108 | 0.062 | 0.039 | -0.011 |
|---|
| PSMD14 |   | 2 | 27 | 36 | 41 | 0.127 | 0.104 | -0.014 | -0.129 | 0.224 |
|---|
| MRE11A |   | 1 | 59 | 202 | 202 | -0.015 | 0.019 | 0.093 | 0.121 | 0.078 |
|---|
| BGN |   | 1 | 59 | 202 | 202 | 0.206 | -0.139 | -0.001 | -0.115 | 0.048 |
|---|
| MMP13 |   | 1 | 59 | 202 | 202 | -0.018 | 0.085 | -0.008 | -0.063 | 0.095 |
|---|
| NCAM1 |   | 1 | 59 | 202 | 202 | -0.052 | 0.167 | 0.010 | 0.056 | -0.074 |
|---|
| LUC7L2 |   | 2 | 27 | 80 | 87 | 0.065 | 0.117 | -0.095 | -0.057 | 0.224 |
|---|
| CFL1 |   | 1 | 59 | 202 | 202 | 0.221 | 0.058 | 0.163 | 0.166 | 0.143 |
|---|
| CKS2 |   | 1 | 59 | 202 | 202 | 0.074 | 0.115 | 0.160 | 0.241 | 0.225 |
|---|
| SSH3 |   | 1 | 59 | 202 | 202 | 0.167 | 0.054 | 0.031 | -0.038 | 0.147 |
|---|
| NAA38 |   | 2 | 27 | 132 | 132 | -0.103 | 0.120 | 0.141 | 0.164 | 0.149 |
|---|
| WRN |   | 1 | 59 | 202 | 202 | 0.007 | 0.159 | 0.047 | -0.045 | 0.142 |
|---|
| MEN1 |   | 1 | 59 | 202 | 202 | 0.158 | -0.078 | -0.058 | 0.097 | -0.039 |
|---|
| NGFR |   | 1 | 59 | 202 | 202 | -0.089 | -0.125 | -0.001 | -0.078 | -0.034 |
|---|
| CDK1 |   | 3 | 13 | 1 | 5 | 0.047 | 0.152 | 0.200 | 0.204 | 0.254 |
|---|
| FGFR1 |   | 1 | 59 | 202 | 202 | 0.130 | 0.112 | 0.058 | -0.094 | -0.002 |
|---|
| MUC1 |   | 1 | 59 | 202 | 202 | -0.039 | 0.062 | -0.049 | -0.020 | 0.014 |
|---|
| ITGB3BP |   | 2 | 27 | 1 | 10 | -0.116 | 0.163 | 0.090 | -0.039 | 0.054 |
|---|
| DCLK1 |   | 1 | 59 | 202 | 202 | 0.075 | -0.055 | 0.077 | -0.017 | 0.056 |
|---|
| PSMC6 |   | 2 | 27 | 36 | 41 | -0.140 | 0.146 | -0.021 | 0.167 | 0.233 |
|---|
| ALB |   | 5 | 4 | 80 | 81 | 0.012 | -0.085 | 0.115 | -0.022 | -0.027 |
|---|
| KRT1 |   | 1 | 59 | 202 | 202 | -0.048 | 0.143 | -0.077 | 0.000 | -0.056 |
|---|
| LUC7L3 |   | 2 | 27 | 132 | 132 | -0.052 | 0.158 | 0.028 | 0.156 | -0.044 |
|---|
| NLRP2 |   | 1 | 59 | 202 | 202 | 0.135 | -0.055 | -0.045 | 0.078 | 0.065 |
|---|
| RAD50 |   | 1 | 59 | 202 | 202 | -0.060 | 0.144 | 0.084 | 0.056 | 0.006 |
|---|
| YWHAZ |   | 1 | 59 | 202 | 202 | 0.122 | -0.011 | 0.121 | 0.048 | 0.108 |
|---|
| ATG5 |   | 1 | 59 | 202 | 202 | -0.093 | 0.077 | 0.014 | -0.011 | 0.120 |
|---|
| MAPK1 |   | 5 | 4 | 110 | 110 | 0.157 | -0.023 | -0.125 | -0.115 | 0.092 |
|---|
| ESR1 |   | 6 | 2 | 1 | 1 | 0.125 | 0.032 | 0.114 | -0.064 | 0.048 |
|---|
| CDKN2A |   | 1 | 59 | 202 | 202 | -0.024 | 0.028 | -0.094 | 0.024 | -0.050 |
|---|
GO Enrichment output for subnetwork 63 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 5.017E-09 | 1.154E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.499E-08 | 1.723E-05 |
|---|
| organ growth | GO:0035265 |  | 3.481E-08 | 2.669E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 3.481E-08 | 2.002E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.932E-08 | 3.189E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 6.932E-08 | 2.657E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.403E-07 | 1.447E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.311E-06 | 3.768E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.726E-06 | 4.412E-04 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.801E-06 | 4.143E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 1.801E-06 | 3.766E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.37E-08 | 3.346E-05 |
|---|
| organ growth | GO:0035265 |  | 1.37E-08 | 1.673E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.37E-08 | 1.115E-05 |
|---|
| dendrite morphogenesis | GO:0048813 |  | 2.729E-08 | 1.667E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.729E-08 | 1.334E-05 |
|---|
| axon extension | GO:0048675 |  | 4.896E-08 | 1.993E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 4.896E-08 | 1.709E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 8.131E-08 | 2.483E-05 |
|---|
| central nervous system neuron axonogenesis | GO:0021955 |  | 8.131E-08 | 2.207E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.273E-07 | 5.553E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 8.516E-07 | 1.891E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.687E-08 | 4.058E-05 |
|---|
| organ growth | GO:0035265 |  | 1.687E-08 | 2.029E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.687E-08 | 1.353E-05 |
|---|
| dendrite morphogenesis | GO:0048813 |  | 3.361E-08 | 2.022E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 3.361E-08 | 1.617E-05 |
|---|
| axon extension | GO:0048675 |  | 6.028E-08 | 2.417E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 6.028E-08 | 2.072E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.001E-07 | 3.01E-05 |
|---|
| central nervous system neuron axonogenesis | GO:0021955 |  | 1.001E-07 | 2.676E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.066E-07 | 7.376E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.665E-07 | 2.114E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 5.017E-09 | 1.154E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.499E-08 | 1.723E-05 |
|---|
| organ growth | GO:0035265 |  | 3.481E-08 | 2.669E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 3.481E-08 | 2.002E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.932E-08 | 3.189E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 6.932E-08 | 2.657E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.403E-07 | 1.447E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.311E-06 | 3.768E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.726E-06 | 4.412E-04 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.801E-06 | 4.143E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 1.801E-06 | 3.766E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 5.017E-09 | 1.154E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.499E-08 | 1.723E-05 |
|---|
| organ growth | GO:0035265 |  | 3.481E-08 | 2.669E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 3.481E-08 | 2.002E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.932E-08 | 3.189E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 6.932E-08 | 2.657E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.403E-07 | 1.447E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.311E-06 | 3.768E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.726E-06 | 4.412E-04 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.801E-06 | 4.143E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 1.801E-06 | 3.766E-04 |
|---|


