Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 999
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3378 | 1.228e-03 | 1.900e-05 | 1.485e-01 | 3.465e-09 |
|---|
| IPC | 0.3497 | 3.342e-01 | 1.659e-01 | 9.326e-01 | 5.171e-02 |
|---|
| Loi | 0.4573 | 1.310e-04 | 0.000e+00 | 6.371e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5865 | 2.000e-06 | 0.000e+00 | 1.427e-01 | 0.000e+00 |
|---|
| Wang | 0.2541 | 8.678e-03 | 7.178e-02 | 2.437e-01 | 1.518e-04 |
|---|
Expression data for subnetwork 999 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
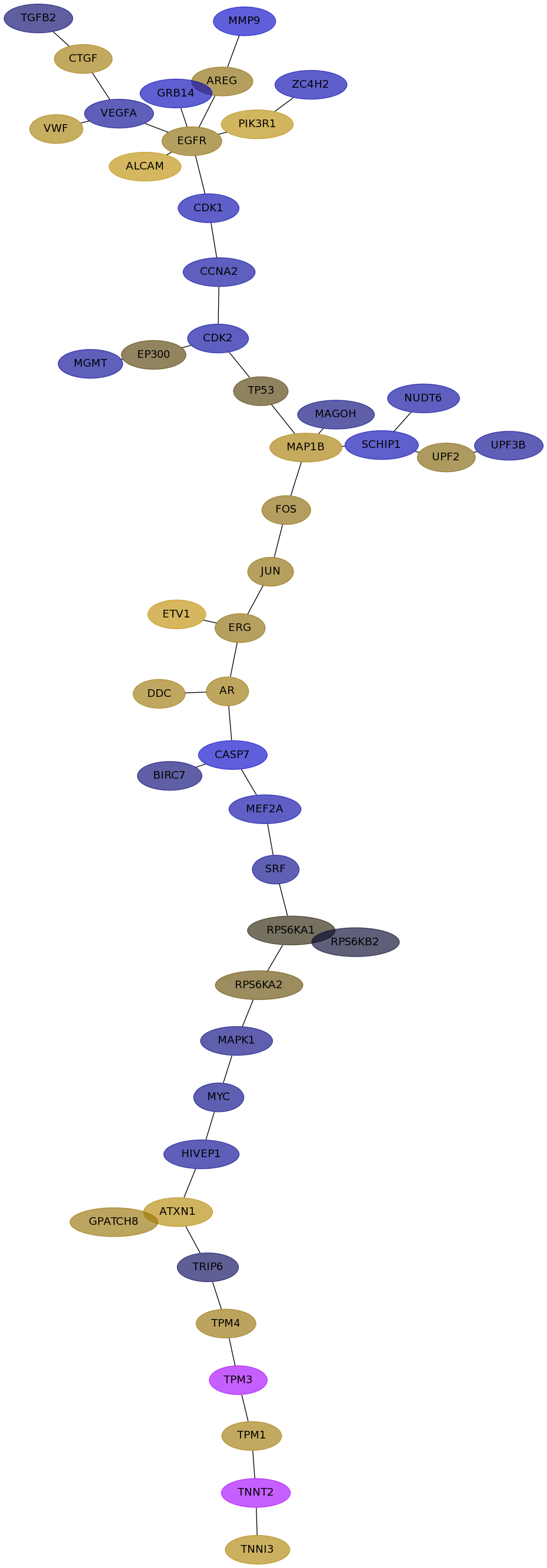
Score for each gene in subnetwork 999 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MMP9 |   | 1 | 652 | 1276 | 1280 | -0.258 | 0.023 | -0.137 | 0.020 | 0.077 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| NUDT6 |   | 1 | 652 | 1276 | 1280 | -0.098 | 0.181 | -0.165 | 0.195 | 0.149 |
|---|
| TNNI3 |   | 1 | 652 | 1276 | 1280 | 0.149 | 0.224 | -0.064 | 0.161 | 0.046 |
|---|
| TRIP6 |   | 3 | 265 | 364 | 362 | -0.020 | 0.170 | 0.145 | 0.034 | -0.213 |
|---|
| UPF3B |   | 1 | 652 | 1276 | 1280 | -0.072 | 0.049 | 0.109 | 0.193 | 0.176 |
|---|
| GRB14 |   | 4 | 198 | 977 | 965 | -0.189 | 0.113 | -0.129 | 0.143 | 0.138 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| MEF2A |   | 5 | 148 | 283 | 287 | -0.123 | 0.022 | 0.036 | 0.265 | 0.124 |
|---|
| TGFB2 |   | 7 | 100 | 57 | 60 | -0.028 | -0.063 | 0.244 | 0.226 | 0.015 |
|---|
| JUN |   | 3 | 265 | 283 | 292 | 0.067 | 0.200 | 0.051 | 0.220 | 0.200 |
|---|
| RPS6KA2 |   | 1 | 652 | 1276 | 1280 | 0.025 | -0.086 | 0.012 | 0.033 | -0.042 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| BIRC7 |   | 1 | 652 | 1276 | 1280 | -0.035 | 0.026 | -0.005 | 0.210 | -0.211 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| GPATCH8 |   | 1 | 652 | 1276 | 1280 | 0.085 | -0.143 | 0.014 | 0.019 | 0.061 |
|---|
| SCHIP1 |   | 2 | 391 | 1039 | 1033 | -0.160 | 0.130 | 0.136 | 0.105 | 0.198 |
|---|
| UPF2 |   | 1 | 652 | 1276 | 1280 | 0.048 | 0.042 | -0.270 | 0.111 | 0.059 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| DDC |   | 4 | 198 | 715 | 700 | 0.093 | -0.121 | 0.157 | -0.078 | 0.114 |
|---|
| RPS6KB2 |   | 2 | 391 | 937 | 928 | -0.006 | 0.033 | -0.161 | 0.131 | -0.000 |
|---|
| ERG |   | 2 | 391 | 444 | 458 | 0.065 | -0.134 | -0.026 | 0.235 | 0.182 |
|---|
| TPM3 |   | 1 | 652 | 1276 | 1280 | undef | 0.135 | 0.036 | undef | undef |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TPM4 |   | 1 | 652 | 1276 | 1280 | 0.081 | -0.048 | 0.161 | 0.034 | -0.078 |
|---|
| MAGOH |   | 1 | 652 | 1276 | 1280 | -0.040 | 0.144 | -0.199 | 0.089 | 0.228 |
|---|
| ATXN1 |   | 6 | 120 | 364 | 355 | 0.171 | 0.026 | 0.192 | 0.175 | 0.163 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| ALCAM |   | 4 | 198 | 1276 | 1276 | 0.206 | -0.208 | 0.193 | 0.067 | -0.010 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| MGMT |   | 2 | 391 | 832 | 831 | -0.082 | 0.184 | -0.022 | 0.201 | 0.196 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| AREG |   | 1 | 652 | 1276 | 1280 | 0.063 | -0.067 | 0.112 | 0.076 | -0.054 |
|---|
| VWF |   | 4 | 198 | 57 | 68 | 0.126 | -0.091 | 0.072 | 0.152 | 0.100 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| SRF |   | 3 | 265 | 283 | 292 | -0.061 | -0.017 | -0.023 | 0.309 | 0.108 |
|---|
| TNNT2 |   | 1 | 652 | 1276 | 1280 | undef | 0.135 | undef | undef | undef |
|---|
| TPM1 |   | 1 | 652 | 1276 | 1280 | 0.103 | -0.014 | 0.228 | 0.253 | 0.131 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| MAPK1 |   | 1 | 652 | 1276 | 1280 | -0.047 | -0.072 | 0.006 | 0.118 | 0.043 |
|---|
| HIVEP1 |   | 6 | 120 | 419 | 410 | -0.072 | -0.199 | 0.069 | 0.000 | -0.042 |
|---|
GO Enrichment output for subnetwork 999 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear-transcribed mRNA catabolic process. nonsense-mediated decay | GO:0000184 |  | 8.454E-08 | 1.945E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.447E-07 | 1.665E-04 |
|---|
| nuclear-transcribed mRNA catabolic process | GO:0000956 |  | 1.926E-07 | 1.477E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.561E-07 | 1.472E-04 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 4.56E-07 | 2.098E-04 |
|---|
| mRNA catabolic process | GO:0006402 |  | 5.333E-07 | 2.044E-04 |
|---|
| RNA catabolic process | GO:0006401 |  | 2.248E-06 | 7.385E-04 |
|---|
| heart morphogenesis | GO:0003007 |  | 3.115E-06 | 8.956E-04 |
|---|
| catechol metabolic process | GO:0009712 |  | 7.628E-06 | 1.949E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 8.867E-06 | 2.039E-03 |
|---|
| positive regulation of heart contraction | GO:0045823 |  | 1.209E-05 | 2.528E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| heart morphogenesis | GO:0003007 |  | 5.774E-08 | 1.41E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.421E-08 | 7.843E-05 |
|---|
| nuclear-transcribed mRNA catabolic process. nonsense-mediated decay | GO:0000184 |  | 7.136E-08 | 5.811E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.284E-07 | 7.841E-05 |
|---|
| nuclear-transcribed mRNA catabolic process | GO:0000956 |  | 1.378E-07 | 6.731E-05 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 1.513E-07 | 6.161E-05 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.856E-07 | 6.476E-05 |
|---|
| regulation of ATPase activity | GO:0043462 |  | 2.736E-07 | 8.356E-05 |
|---|
| mRNA catabolic process | GO:0006402 |  | 3.636E-07 | 9.871E-05 |
|---|
| ventricular cardiac muscle morphogenesis | GO:0055010 |  | 4.575E-07 | 1.118E-04 |
|---|
| cardiac muscle tissue morphogenesis | GO:0055008 |  | 1.306E-06 | 2.9E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear-transcribed mRNA catabolic process. nonsense-mediated decay | GO:0000184 |  | 5.704E-08 | 1.372E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.81E-08 | 8.192E-05 |
|---|
| nuclear-transcribed mRNA catabolic process | GO:0000956 |  | 1.155E-07 | 9.264E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.378E-07 | 8.287E-05 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 1.7E-07 | 8.181E-05 |
|---|
| mRNA catabolic process | GO:0006402 |  | 3.235E-07 | 1.297E-04 |
|---|
| RNA catabolic process | GO:0006401 |  | 1.548E-06 | 5.32E-04 |
|---|
| heart morphogenesis | GO:0003007 |  | 1.865E-06 | 5.61E-04 |
|---|
| catechol metabolic process | GO:0009712 |  | 3.866E-06 | 1.033E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 4.448E-06 | 1.07E-03 |
|---|
| positive regulation of heart contraction | GO:0045823 |  | 5.738E-06 | 1.255E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear-transcribed mRNA catabolic process. nonsense-mediated decay | GO:0000184 |  | 8.454E-08 | 1.945E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.447E-07 | 1.665E-04 |
|---|
| nuclear-transcribed mRNA catabolic process | GO:0000956 |  | 1.926E-07 | 1.477E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.561E-07 | 1.472E-04 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 4.56E-07 | 2.098E-04 |
|---|
| mRNA catabolic process | GO:0006402 |  | 5.333E-07 | 2.044E-04 |
|---|
| RNA catabolic process | GO:0006401 |  | 2.248E-06 | 7.385E-04 |
|---|
| heart morphogenesis | GO:0003007 |  | 3.115E-06 | 8.956E-04 |
|---|
| catechol metabolic process | GO:0009712 |  | 7.628E-06 | 1.949E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 8.867E-06 | 2.039E-03 |
|---|
| positive regulation of heart contraction | GO:0045823 |  | 1.209E-05 | 2.528E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear-transcribed mRNA catabolic process. nonsense-mediated decay | GO:0000184 |  | 8.454E-08 | 1.945E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.447E-07 | 1.665E-04 |
|---|
| nuclear-transcribed mRNA catabolic process | GO:0000956 |  | 1.926E-07 | 1.477E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.561E-07 | 1.472E-04 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 4.56E-07 | 2.098E-04 |
|---|
| mRNA catabolic process | GO:0006402 |  | 5.333E-07 | 2.044E-04 |
|---|
| RNA catabolic process | GO:0006401 |  | 2.248E-06 | 7.385E-04 |
|---|
| heart morphogenesis | GO:0003007 |  | 3.115E-06 | 8.956E-04 |
|---|
| catechol metabolic process | GO:0009712 |  | 7.628E-06 | 1.949E-03 |
|---|
| phenol metabolic process | GO:0018958 |  | 8.867E-06 | 2.039E-03 |
|---|
| positive regulation of heart contraction | GO:0045823 |  | 1.209E-05 | 2.528E-03 |
|---|


