Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 967
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3360 | 1.337e-03 | 2.200e-05 | 1.541e-01 | 4.533e-09 |
|---|
| IPC | 0.3676 | 3.048e-01 | 1.418e-01 | 9.264e-01 | 4.003e-02 |
|---|
| Loi | 0.4538 | 1.540e-04 | 0.000e+00 | 6.576e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5862 | 2.000e-06 | 0.000e+00 | 1.435e-01 | 0.000e+00 |
|---|
| Wang | 0.2546 | 8.508e-03 | 7.090e-02 | 2.415e-01 | 1.457e-04 |
|---|
Expression data for subnetwork 967 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
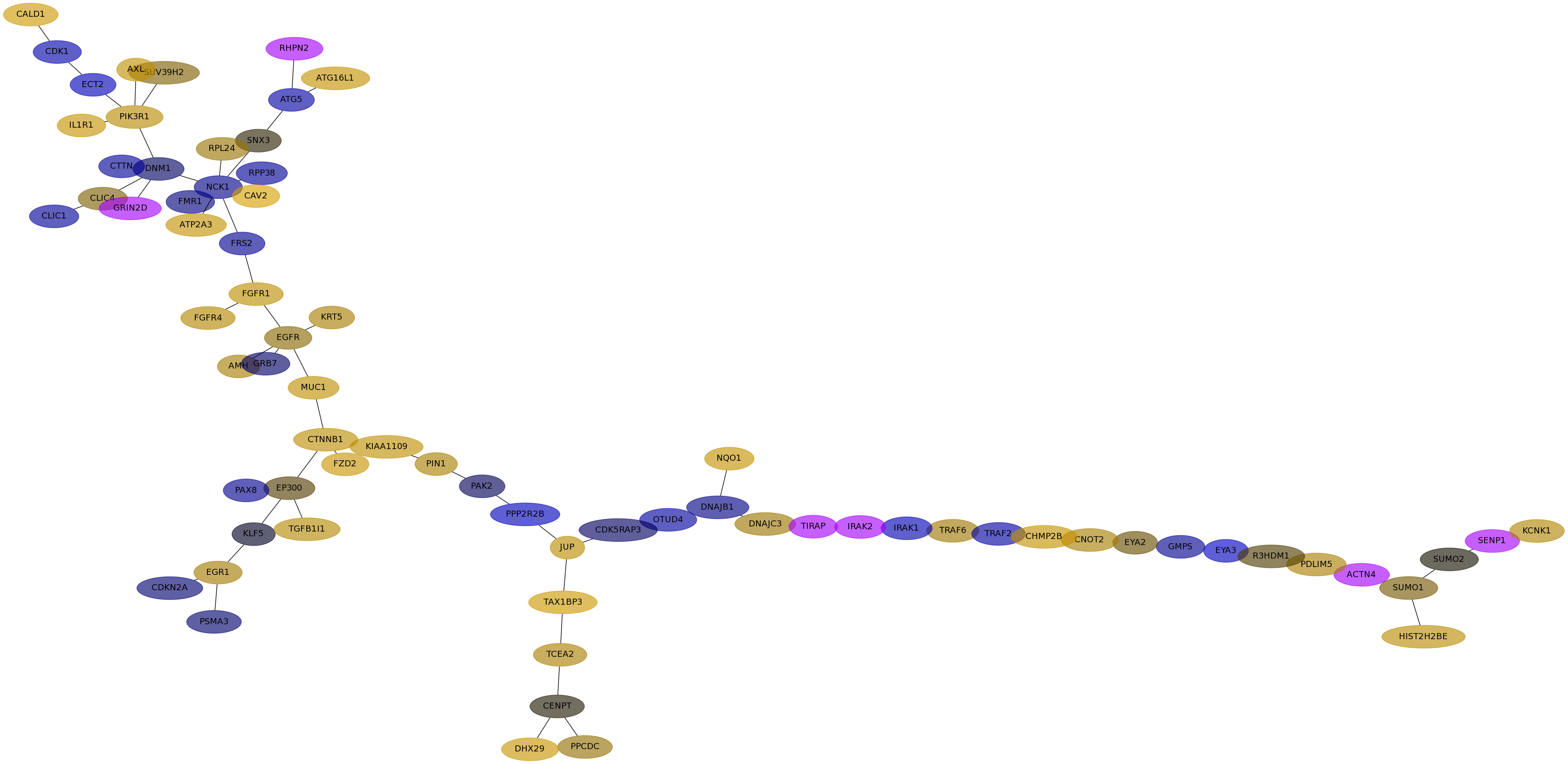
Score for each gene in subnetwork 967 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TRAF6 |   | 2 | 391 | 1106 | 1100 | 0.102 | -0.060 | -0.033 | 0.073 | -0.048 |
|---|
| PSMA3 |   | 2 | 391 | 444 | 458 | -0.033 | 0.116 | -0.149 | -0.008 | 0.014 |
|---|
| NQO1 |   | 6 | 120 | 536 | 535 | 0.243 | -0.115 | 0.069 | -0.189 | 0.118 |
|---|
| DNAJC3 |   | 1 | 652 | 1106 | 1110 | 0.096 | -0.039 | -0.065 | -0.040 | -0.173 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| HIST2H2BE |   | 5 | 148 | 492 | 498 | 0.196 | -0.086 | 0.123 | -0.180 | -0.104 |
|---|
| RHPN2 |   | 1 | 652 | 1106 | 1110 | undef | 0.274 | 0.047 | undef | undef |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| PPCDC |   | 1 | 652 | 1106 | 1110 | 0.080 | 0.248 | -0.056 | 0.222 | -0.085 |
|---|
| TAX1BP3 |   | 5 | 148 | 552 | 541 | 0.292 | 0.150 | 0.219 | 0.070 | 0.206 |
|---|
| CLIC1 |   | 1 | 652 | 1106 | 1110 | -0.092 | 0.208 | 0.060 | -0.019 | 0.178 |
|---|
| CTTN |   | 1 | 652 | 1106 | 1110 | -0.086 | -0.063 | 0.232 | -0.064 | 0.151 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| FRS2 |   | 1 | 652 | 1106 | 1110 | -0.077 | 0.252 | 0.093 | 0.174 | -0.095 |
|---|
| SENP1 |   | 1 | 652 | 1106 | 1110 | undef | 0.126 | -0.034 | undef | undef |
|---|
| KRT5 |   | 5 | 148 | 923 | 898 | 0.129 | -0.125 | 0.046 | 0.064 | -0.190 |
|---|
| RPP38 |   | 1 | 652 | 1106 | 1110 | -0.086 | 0.104 | -0.281 | -0.121 | 0.046 |
|---|
| NCK1 |   | 3 | 265 | 206 | 208 | -0.065 | 0.135 | -0.100 | -0.059 | 0.151 |
|---|
| ATG16L1 |   | 2 | 391 | 756 | 756 | 0.244 | -0.123 | 0.108 | -0.112 | -0.175 |
|---|
| EYA3 |   | 1 | 652 | 1106 | 1110 | -0.228 | -0.034 | 0.053 | -0.020 | -0.142 |
|---|
| ECT2 |   | 2 | 391 | 910 | 899 | -0.195 | 0.223 | -0.045 | 0.040 | -0.058 |
|---|
| R3HDM1 |   | 1 | 652 | 1106 | 1110 | 0.016 | -0.108 | -0.192 | -0.049 | 0.223 |
|---|
| CHMP2B |   | 3 | 265 | 419 | 419 | 0.250 | 0.128 | 0.134 | -0.039 | 0.067 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| CDK5RAP3 |   | 1 | 652 | 1106 | 1110 | -0.025 | -0.018 | 0.051 | -0.127 | -0.084 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| KCNK1 |   | 1 | 652 | 1106 | 1110 | 0.158 | 0.119 | 0.214 | -0.329 | 0.037 |
|---|
| AXL |   | 1 | 652 | 1106 | 1110 | 0.224 | -0.043 | 0.132 | -0.044 | -0.013 |
|---|
| ACTN4 |   | 6 | 120 | 859 | 834 | undef | 0.035 | 0.180 | undef | undef |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| ATG5 |   | 2 | 391 | 756 | 756 | -0.116 | 0.195 | -0.113 | -0.013 | 0.210 |
|---|
| EYA2 |   | 5 | 148 | 419 | 415 | 0.028 | -0.034 | 0.113 | -0.142 | -0.019 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| JUP |   | 1 | 652 | 1106 | 1110 | 0.202 | -0.042 | 0.182 | -0.155 | -0.034 |
|---|
| TRAF2 |   | 6 | 120 | 206 | 200 | -0.107 | 0.185 | -0.172 | -0.013 | -0.092 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| DNAJB1 |   | 1 | 652 | 1106 | 1110 | -0.058 | -0.007 | 0.052 | 0.039 | -0.007 |
|---|
| SUMO1 |   | 3 | 265 | 1106 | 1083 | 0.038 | 0.245 | -0.007 | 0.144 | 0.118 |
|---|
| CLIC4 |   | 2 | 391 | 1106 | 1100 | 0.048 | 0.141 | 0.196 | 0.282 | 0.106 |
|---|
| SUV39H2 |   | 3 | 265 | 859 | 843 | 0.049 | 0.150 | -0.097 | -0.057 | 0.028 |
|---|
| IRAK1 |   | 4 | 198 | 206 | 205 | -0.170 | 0.266 | -0.023 | 0.092 | -0.102 |
|---|
| CALD1 |   | 1 | 652 | 1106 | 1110 | 0.307 | -0.209 | 0.242 | 0.032 | 0.229 |
|---|
| AMH |   | 3 | 265 | 778 | 770 | 0.124 | 0.103 | 0.056 | 0.113 | 0.301 |
|---|
| SNX3 |   | 1 | 652 | 1106 | 1110 | 0.006 | 0.147 | -0.079 | 0.085 | 0.215 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| GMPS |   | 1 | 652 | 1106 | 1110 | -0.075 | 0.083 | -0.022 | 0.044 | 0.151 |
|---|
| FMR1 |   | 3 | 265 | 679 | 667 | -0.047 | 0.137 | -0.104 | 0.352 | 0.113 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| DNM1 |   | 1 | 652 | 1106 | 1110 | -0.025 | -0.203 | 0.095 | 0.163 | 0.082 |
|---|
| DHX29 |   | 1 | 652 | 1106 | 1110 | 0.269 | 0.147 | 0.275 | -0.129 | 0.030 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| KIAA1109 |   | 2 | 391 | 1106 | 1100 | 0.220 | -0.036 | 0.201 | 0.065 | 0.173 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| IL1R1 |   | 8 | 84 | 536 | 515 | 0.257 | -0.211 | 0.178 | -0.071 | 0.003 |
|---|
| CNOT2 |   | 3 | 265 | 419 | 419 | 0.132 | 0.202 | 0.114 | 0.112 | 0.123 |
|---|
| TCEA2 |   | 1 | 652 | 1106 | 1110 | 0.137 | 0.115 | 0.007 | 0.197 | -0.020 |
|---|
| PAX8 |   | 4 | 198 | 778 | 766 | -0.072 | 0.008 | -0.083 | 0.032 | -0.002 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| IRAK2 |   | 3 | 265 | 1094 | 1070 | undef | 0.183 | 0.023 | undef | undef |
|---|
| OTUD4 |   | 2 | 391 | 900 | 897 | -0.107 | -0.204 | -0.012 | 0.227 | -0.092 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| RPL24 |   | 1 | 652 | 1106 | 1110 | 0.094 | 0.160 | 0.046 | 0.051 | 0.055 |
|---|
| CENPT |   | 1 | 652 | 1106 | 1110 | 0.004 | -0.125 | 0.111 | -0.224 | 0.070 |
|---|
| PDLIM5 |   | 1 | 652 | 1106 | 1110 | 0.136 | -0.091 | 0.242 | -0.085 | 0.117 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| TIRAP |   | 1 | 652 | 1106 | 1110 | undef | 0.011 | 0.004 | undef | undef |
|---|
GO Enrichment output for subnetwork 967 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 4.844E-11 | 1.114E-07 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 3.881E-10 | 4.463E-07 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 2.148E-06 | 1.647E-03 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 3.706E-06 | 2.131E-03 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 4.46E-06 | 2.052E-03 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 4.968E-06 | 1.904E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 6.075E-06 | 1.996E-03 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 7.803E-06 | 2.243E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 9.466E-06 | 2.419E-03 |
|---|
| macroautophagy | GO:0016236 |  | 9.878E-06 | 2.272E-03 |
|---|
| positive regulation of cytokine production during immune response | GO:0002720 |  | 9.878E-06 | 2.065E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 6.639E-12 | 1.622E-08 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 5.65E-10 | 6.901E-07 |
|---|
| I-kappaB kinase/NF-kappaB cascade | GO:0007249 |  | 5.916E-07 | 4.818E-04 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 7.338E-07 | 4.482E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.67E-06 | 8.159E-04 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.888E-06 | 7.687E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.319E-06 | 8.094E-04 |
|---|
| negative regulation of binding | GO:0051100 |  | 2.844E-06 | 8.684E-04 |
|---|
| positive regulation of cytokine production during immune response | GO:0002720 |  | 3.625E-06 | 9.84E-04 |
|---|
| regulation of transcription factor activity | GO:0051090 |  | 4.675E-06 | 1.142E-03 |
|---|
| positive regulation of production of molecular mediator of immune response | GO:0002702 |  | 1.006E-05 | 2.235E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 9.54E-12 | 2.295E-08 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 5.427E-10 | 6.529E-07 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 1.172E-06 | 9.4E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.951E-06 | 1.173E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 2.919E-06 | 1.405E-03 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.951E-06 | 1.183E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 4.342E-06 | 1.493E-03 |
|---|
| positive regulation of cytokine production during immune response | GO:0002720 |  | 4.355E-06 | 1.31E-03 |
|---|
| regulation of transcription factor activity | GO:0051090 |  | 6.096E-06 | 1.63E-03 |
|---|
| positive regulation of production of molecular mediator of immune response | GO:0002702 |  | 1.208E-05 | 2.908E-03 |
|---|
| I-kappaB kinase/NF-kappaB cascade | GO:0007249 |  | 1.469E-05 | 3.212E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 4.844E-11 | 1.114E-07 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 3.881E-10 | 4.463E-07 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 2.148E-06 | 1.647E-03 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 3.706E-06 | 2.131E-03 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 4.46E-06 | 2.052E-03 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 4.968E-06 | 1.904E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 6.075E-06 | 1.996E-03 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 7.803E-06 | 2.243E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 9.466E-06 | 2.419E-03 |
|---|
| macroautophagy | GO:0016236 |  | 9.878E-06 | 2.272E-03 |
|---|
| positive regulation of cytokine production during immune response | GO:0002720 |  | 9.878E-06 | 2.065E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 4.844E-11 | 1.114E-07 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 3.881E-10 | 4.463E-07 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 2.148E-06 | 1.647E-03 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 3.706E-06 | 2.131E-03 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 4.46E-06 | 2.052E-03 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 4.968E-06 | 1.904E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 6.075E-06 | 1.996E-03 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 7.803E-06 | 2.243E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 9.466E-06 | 2.419E-03 |
|---|
| macroautophagy | GO:0016236 |  | 9.878E-06 | 2.272E-03 |
|---|
| positive regulation of cytokine production during immune response | GO:0002720 |  | 9.878E-06 | 2.065E-03 |
|---|


