Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 847
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3448 | 8.880e-04 | 1.100e-05 | 1.288e-01 | 1.258e-09 |
|---|
| IPC | 0.3310 | 3.650e-01 | 1.928e-01 | 9.390e-01 | 6.609e-02 |
|---|
| Loi | 0.4473 | 2.030e-04 | 0.000e+00 | 6.963e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5656 | 5.000e-06 | 0.000e+00 | 1.987e-01 | 0.000e+00 |
|---|
| Wang | 0.2410 | 1.434e-02 | 9.804e-02 | 3.052e-01 | 4.291e-04 |
|---|
Expression data for subnetwork 847 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
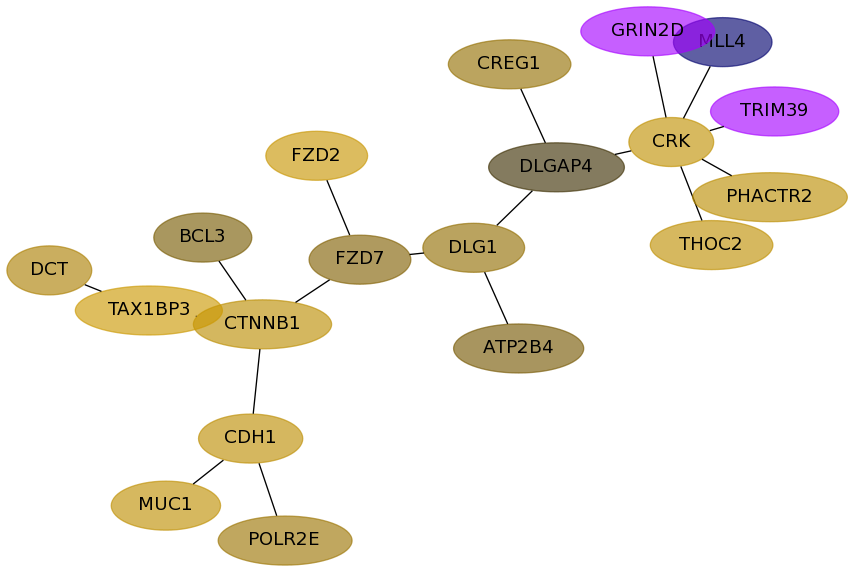
Score for each gene in subnetwork 847 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CREG1 |   | 5 | 148 | 691 | 670 | 0.080 | 0.029 | 0.018 | -0.189 | -0.176 |
|---|
| DLGAP4 |   | 5 | 148 | 326 | 330 | 0.010 | -0.029 | 0.180 | -0.201 | -0.043 |
|---|
| PHACTR2 |   | 5 | 148 | 271 | 267 | 0.203 | -0.057 | -0.051 | 0.143 | 0.164 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| DCT |   | 1 | 652 | 1458 | 1458 | 0.137 | undef | -0.019 | 0.000 | -0.092 |
|---|
| FZD7 |   | 5 | 148 | 552 | 541 | 0.051 | -0.219 | 0.062 | 0.062 | -0.059 |
|---|
| TRIM39 |   | 2 | 391 | 412 | 423 | undef | -0.144 | 0.092 | undef | undef |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| DLG1 |   | 1 | 652 | 1458 | 1458 | 0.076 | -0.016 | 0.173 | 0.244 | 0.123 |
|---|
| POLR2E |   | 4 | 198 | 598 | 577 | 0.095 | 0.127 | 0.039 | 0.166 | -0.018 |
|---|
| MLL4 |   | 4 | 198 | 412 | 409 | -0.032 | -0.093 | 0.052 | -0.101 | -0.153 |
|---|
| TAX1BP3 |   | 5 | 148 | 552 | 541 | 0.292 | 0.150 | 0.219 | 0.070 | 0.206 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| BCL3 |   | 2 | 391 | 679 | 678 | 0.042 | -0.025 | 0.175 | -0.150 | -0.048 |
|---|
| ATP2B4 |   | 1 | 652 | 1458 | 1458 | 0.038 | 0.142 | 0.177 | -0.004 | 0.070 |
|---|
GO Enrichment output for subnetwork 847 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of transcription factor import into nucleus | GO:0042990 |  | 1.381E-05 | 0.03175375 |
|---|
| regulation of protein import into nucleus | GO:0042306 |  | 2.824E-05 | 0.0324808 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.984E-05 | 0.02287991 |
|---|
| response to protozoan | GO:0001562 |  | 4.107E-05 | 0.02361515 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 4.107E-05 | 0.01889212 |
|---|
| response to UV-C | GO:0010225 |  | 4.107E-05 | 0.01574343 |
|---|
| CD4-positive. alpha beta T cell differentiation | GO:0043367 |  | 4.107E-05 | 0.01349437 |
|---|
| lymphocyte activation during immune response | GO:0002285 |  | 4.107E-05 | 0.01180757 |
|---|
| regulation of sensory perception | GO:0051931 |  | 4.107E-05 | 0.01049562 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 5.016E-05 | 0.01153747 |
|---|
| regulation of tumor necrosis factor biosynthetic process | GO:0042534 |  | 6.152E-05 | 0.01286429 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.549E-07 | 8.669E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 9.749E-07 | 1.191E-03 |
|---|
| regulation of transcription factor import into nucleus | GO:0042990 |  | 5.514E-06 | 4.491E-03 |
|---|
| regulation of protein import into nucleus | GO:0042306 |  | 1.046E-05 | 6.389E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.631E-05 | 7.97E-03 |
|---|
| response to protozoan | GO:0001562 |  | 1.707E-05 | 6.95E-03 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 1.707E-05 | 5.957E-03 |
|---|
| T cell activation during immune response | GO:0002286 |  | 1.707E-05 | 5.213E-03 |
|---|
| regulation of sensory perception | GO:0051931 |  | 1.707E-05 | 4.634E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 2.23E-05 | 5.447E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.399E-05 | 5.328E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.242E-07 | 1.021E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.599E-06 | 1.924E-03 |
|---|
| regulation of transcription factor import into nucleus | GO:0042990 |  | 9.03E-06 | 7.242E-03 |
|---|
| regulation of protein import into nucleus | GO:0042306 |  | 1.711E-05 | 0.01029422 |
|---|
| response to protozoan | GO:0001562 |  | 2.377E-05 | 0.01143599 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 2.377E-05 | 9.53E-03 |
|---|
| CD4-positive. alpha beta T cell differentiation | GO:0043367 |  | 2.377E-05 | 8.169E-03 |
|---|
| lymphocyte activation during immune response | GO:0002285 |  | 2.377E-05 | 7.147E-03 |
|---|
| regulation of sensory perception | GO:0051931 |  | 2.377E-05 | 6.353E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.453E-05 | 5.902E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 3.379E-05 | 7.391E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of transcription factor import into nucleus | GO:0042990 |  | 1.381E-05 | 0.03175375 |
|---|
| regulation of protein import into nucleus | GO:0042306 |  | 2.824E-05 | 0.0324808 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.984E-05 | 0.02287991 |
|---|
| response to protozoan | GO:0001562 |  | 4.107E-05 | 0.02361515 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 4.107E-05 | 0.01889212 |
|---|
| response to UV-C | GO:0010225 |  | 4.107E-05 | 0.01574343 |
|---|
| CD4-positive. alpha beta T cell differentiation | GO:0043367 |  | 4.107E-05 | 0.01349437 |
|---|
| lymphocyte activation during immune response | GO:0002285 |  | 4.107E-05 | 0.01180757 |
|---|
| regulation of sensory perception | GO:0051931 |  | 4.107E-05 | 0.01049562 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 5.016E-05 | 0.01153747 |
|---|
| regulation of tumor necrosis factor biosynthetic process | GO:0042534 |  | 6.152E-05 | 0.01286429 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of transcription factor import into nucleus | GO:0042990 |  | 1.381E-05 | 0.03175375 |
|---|
| regulation of protein import into nucleus | GO:0042306 |  | 2.824E-05 | 0.0324808 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.984E-05 | 0.02287991 |
|---|
| response to protozoan | GO:0001562 |  | 4.107E-05 | 0.02361515 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 4.107E-05 | 0.01889212 |
|---|
| response to UV-C | GO:0010225 |  | 4.107E-05 | 0.01574343 |
|---|
| CD4-positive. alpha beta T cell differentiation | GO:0043367 |  | 4.107E-05 | 0.01349437 |
|---|
| lymphocyte activation during immune response | GO:0002285 |  | 4.107E-05 | 0.01180757 |
|---|
| regulation of sensory perception | GO:0051931 |  | 4.107E-05 | 0.01049562 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 5.016E-05 | 0.01153747 |
|---|
| regulation of tumor necrosis factor biosynthetic process | GO:0042534 |  | 6.152E-05 | 0.01286429 |
|---|


