Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 811
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3387 | 1.179e-03 | 1.800e-05 | 1.459e-01 | 3.096e-09 |
|---|
| IPC | 0.3333 | 3.611e-01 | 1.893e-01 | 9.382e-01 | 6.413e-02 |
|---|
| Loi | 0.4505 | 1.770e-04 | 0.000e+00 | 6.770e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5654 | 6.000e-06 | 0.000e+00 | 1.993e-01 | 0.000e+00 |
|---|
| Wang | 0.2388 | 1.553e-02 | 1.030e-01 | 3.160e-01 | 5.057e-04 |
|---|
Expression data for subnetwork 811 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
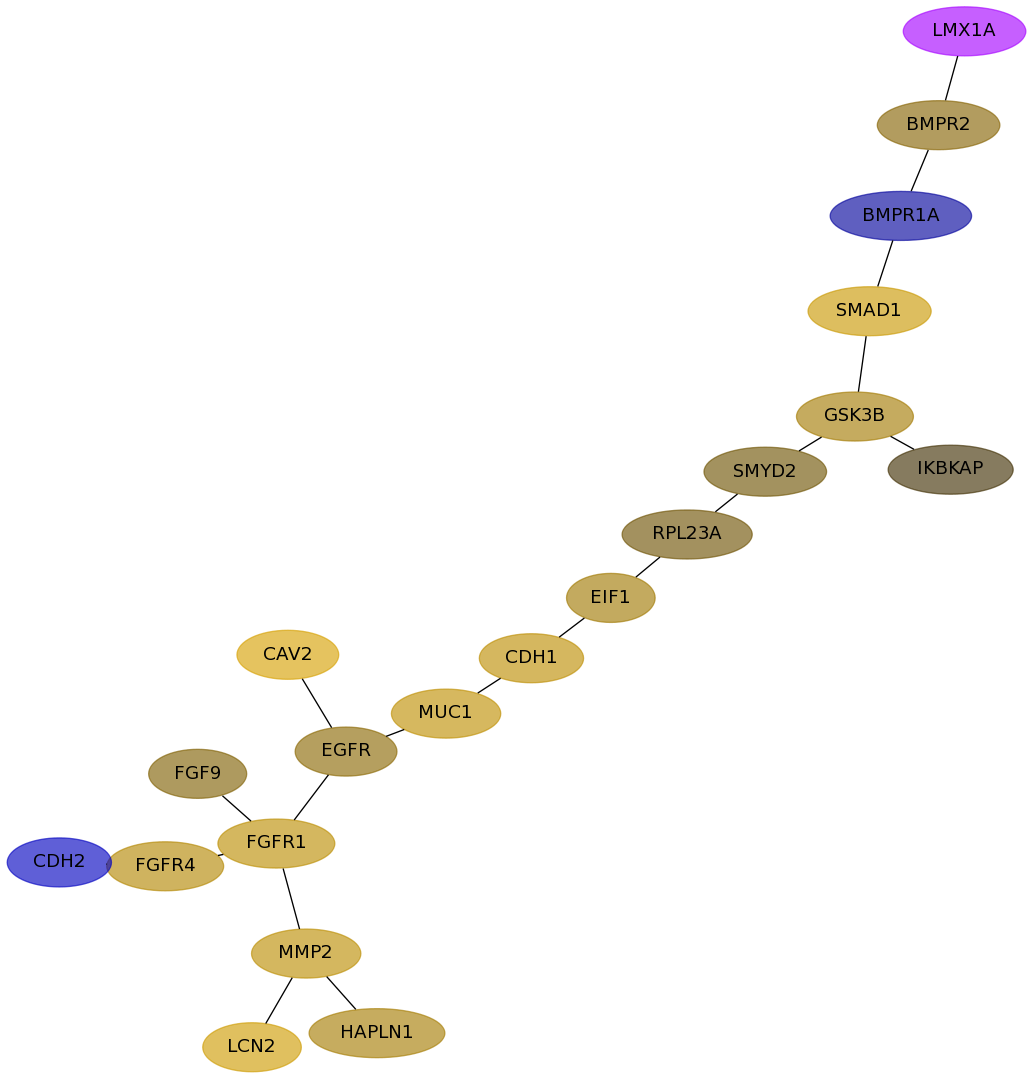
Score for each gene in subnetwork 811 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| FGF9 |   | 5 | 148 | 262 | 265 | 0.050 | -0.096 | 0.087 | 0.014 | 0.174 |
|---|
| BMPR2 |   | 1 | 652 | 1469 | 1469 | 0.056 | -0.132 | 0.081 | -0.029 | -0.021 |
|---|
| LCN2 |   | 6 | 120 | 797 | 779 | 0.316 | 0.113 | 0.145 | -0.098 | -0.284 |
|---|
| IKBKAP |   | 5 | 148 | 364 | 358 | 0.010 | 0.089 | 0.184 | 0.142 | -0.017 |
|---|
| SMYD2 |   | 4 | 198 | 148 | 162 | 0.032 | 0.260 | 0.068 | 0.179 | 0.191 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| HAPLN1 |   | 3 | 265 | 1103 | 1080 | 0.119 | 0.064 | 0.036 | 0.176 | 0.226 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| MMP2 |   | 7 | 100 | 797 | 778 | 0.207 | -0.105 | 0.186 | -0.024 | 0.162 |
|---|
| EIF1 |   | 1 | 652 | 1469 | 1469 | 0.108 | 0.114 | -0.046 | 0.187 | 0.137 |
|---|
| LMX1A |   | 1 | 652 | 1469 | 1469 | undef | undef | 0.306 | undef | undef |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| BMPR1A |   | 2 | 391 | 665 | 660 | -0.096 | -0.108 | 0.122 | 0.140 | -0.064 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| CDH2 |   | 7 | 100 | 33 | 37 | -0.228 | 0.169 | 0.109 | -0.016 | 0.155 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 811 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.197E-07 | 2.753E-04 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 1.494E-07 | 1.718E-04 |
|---|
| lung development | GO:0030324 |  | 3.213E-07 | 2.463E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 3.252E-07 | 1.87E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 4.13E-07 | 1.9E-04 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 4.818E-07 | 1.847E-04 |
|---|
| respiratory system development | GO:0060541 |  | 4.849E-07 | 1.593E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 6.222E-07 | 1.789E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 6.222E-07 | 1.59E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 7.205E-07 | 1.657E-04 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 7.614E-07 | 1.592E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 5.589E-08 | 1.365E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 9.883E-08 | 1.207E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 1.387E-07 | 1.129E-04 |
|---|
| lung development | GO:0030324 |  | 1.659E-07 | 1.013E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 2.057E-07 | 1.005E-04 |
|---|
| respiratory system development | GO:0060541 |  | 2.362E-07 | 9.617E-05 |
|---|
| mesoderm formation | GO:0001707 |  | 3.309E-07 | 1.155E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 3.309E-07 | 1.01E-04 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 3.496E-07 | 9.489E-05 |
|---|
| formation of primary germ layer | GO:0001704 |  | 4.255E-07 | 1.04E-04 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 4.641E-07 | 1.031E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 6.735E-08 | 1.62E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.22E-08 | 9.888E-05 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 1.67E-07 | 1.34E-04 |
|---|
| lung development | GO:0030324 |  | 1.792E-07 | 1.078E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 2.236E-07 | 1.076E-04 |
|---|
| respiratory system development | GO:0060541 |  | 2.577E-07 | 1.033E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 3.491E-07 | 1.2E-04 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 3.575E-07 | 1.075E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 3.984E-07 | 1.065E-04 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 4.11E-07 | 9.888E-05 |
|---|
| formation of primary germ layer | GO:0001704 |  | 4.527E-07 | 9.903E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.197E-07 | 2.753E-04 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 1.494E-07 | 1.718E-04 |
|---|
| lung development | GO:0030324 |  | 3.213E-07 | 2.463E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 3.252E-07 | 1.87E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 4.13E-07 | 1.9E-04 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 4.818E-07 | 1.847E-04 |
|---|
| respiratory system development | GO:0060541 |  | 4.849E-07 | 1.593E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 6.222E-07 | 1.789E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 6.222E-07 | 1.59E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 7.205E-07 | 1.657E-04 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 7.614E-07 | 1.592E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.197E-07 | 2.753E-04 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 1.494E-07 | 1.718E-04 |
|---|
| lung development | GO:0030324 |  | 3.213E-07 | 2.463E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 3.252E-07 | 1.87E-04 |
|---|
| respiratory tube development | GO:0030323 |  | 4.13E-07 | 1.9E-04 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 4.818E-07 | 1.847E-04 |
|---|
| respiratory system development | GO:0060541 |  | 4.849E-07 | 1.593E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 6.222E-07 | 1.789E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 6.222E-07 | 1.59E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 7.205E-07 | 1.657E-04 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 7.614E-07 | 1.592E-04 |
|---|


