Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 799
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3355 | 1.368e-03 | 2.200e-05 | 1.556e-01 | 4.684e-09 |
|---|
| IPC | 0.3355 | 3.576e-01 | 1.862e-01 | 9.375e-01 | 6.241e-02 |
|---|
| Loi | 0.4496 | 1.840e-04 | 0.000e+00 | 6.823e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5700 | 4.000e-06 | 0.000e+00 | 1.861e-01 | 0.000e+00 |
|---|
| Wang | 0.2385 | 1.567e-02 | 1.036e-01 | 3.173e-01 | 5.150e-04 |
|---|
Expression data for subnetwork 799 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
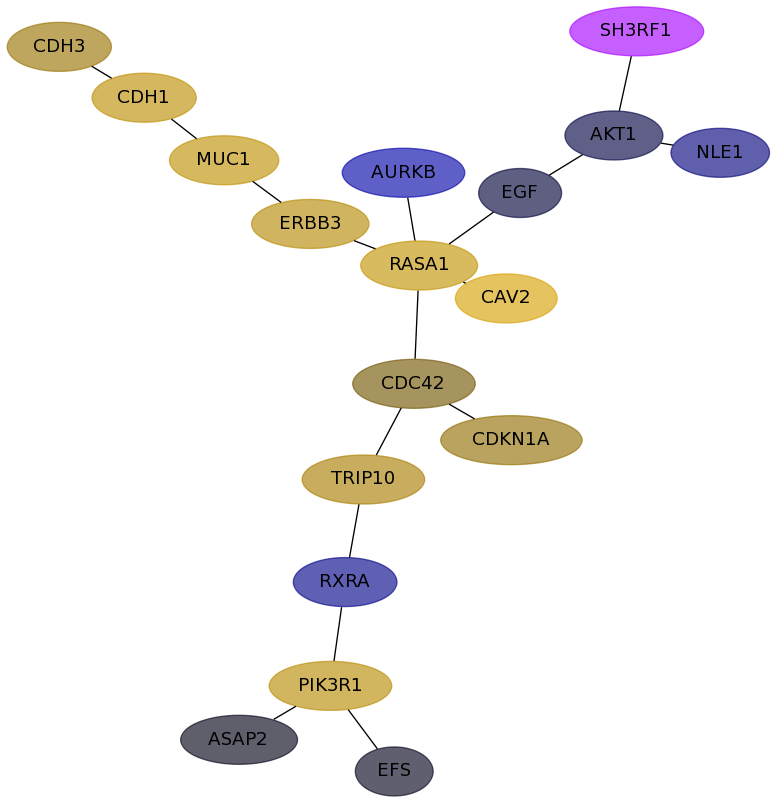
Score for each gene in subnetwork 799 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| TRIP10 |   | 3 | 265 | 715 | 705 | 0.127 | 0.012 | 0.154 | 0.049 | 0.144 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| RXRA |   | 3 | 265 | 283 | 292 | -0.062 | -0.055 | 0.134 | -0.027 | -0.096 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| EFS |   | 4 | 198 | 129 | 130 | -0.003 | -0.024 | 0.300 | -0.003 | -0.034 |
|---|
| AURKB |   | 3 | 265 | 1236 | 1219 | -0.132 | 0.179 | -0.189 | 0.086 | -0.028 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| NLE1 |   | 1 | 652 | 1461 | 1461 | -0.043 | 0.138 | -0.086 | -0.018 | 0.044 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| ASAP2 |   | 1 | 652 | 1461 | 1461 | -0.003 | -0.110 | 0.001 | 0.103 | -0.027 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| ERBB3 |   | 5 | 148 | 206 | 202 | 0.175 | -0.171 | -0.007 | -0.141 | -0.098 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 799 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 3.718E-08 | 8.552E-05 |
|---|
| nuclear migration | GO:0007097 |  | 5.037E-08 | 5.792E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.037E-08 | 3.861E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.006E-07 | 5.785E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.759E-07 | 8.089E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.759E-07 | 6.741E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.81E-07 | 9.234E-05 |
|---|
| nucleus localization | GO:0051647 |  | 4.21E-07 | 1.211E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 6.008E-07 | 1.535E-04 |
|---|
| regulation of cellular component biogenesis | GO:0044087 |  | 6.821E-07 | 1.569E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 8.251E-07 | 1.725E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.359E-08 | 3.321E-05 |
|---|
| nuclear migration | GO:0007097 |  | 3.29E-08 | 4.018E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.29E-08 | 2.679E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.196E-08 | 5.616E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 9.196E-08 | 4.493E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.378E-07 | 5.612E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.378E-07 | 4.81E-05 |
|---|
| nucleus localization | GO:0051647 |  | 2.703E-07 | 8.253E-05 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.6E-07 | 9.773E-05 |
|---|
| filopodium assembly | GO:0046847 |  | 1.108E-06 | 2.707E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.267E-06 | 2.813E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.796E-08 | 4.322E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.329E-08 | 5.207E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.329E-08 | 3.472E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.568E-08 | 4.552E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.568E-08 | 3.642E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.21E-07 | 4.851E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.813E-07 | 6.231E-05 |
|---|
| nucleus localization | GO:0051647 |  | 3.555E-07 | 1.069E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.735E-07 | 1.266E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.201E-06 | 2.889E-04 |
|---|
| microspike assembly | GO:0030035 |  | 1.457E-06 | 3.187E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 3.718E-08 | 8.552E-05 |
|---|
| nuclear migration | GO:0007097 |  | 5.037E-08 | 5.792E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.037E-08 | 3.861E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.006E-07 | 5.785E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.759E-07 | 8.089E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.759E-07 | 6.741E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.81E-07 | 9.234E-05 |
|---|
| nucleus localization | GO:0051647 |  | 4.21E-07 | 1.211E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 6.008E-07 | 1.535E-04 |
|---|
| regulation of cellular component biogenesis | GO:0044087 |  | 6.821E-07 | 1.569E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 8.251E-07 | 1.725E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 3.718E-08 | 8.552E-05 |
|---|
| nuclear migration | GO:0007097 |  | 5.037E-08 | 5.792E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.037E-08 | 3.861E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.006E-07 | 5.785E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.759E-07 | 8.089E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.759E-07 | 6.741E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.81E-07 | 9.234E-05 |
|---|
| nucleus localization | GO:0051647 |  | 4.21E-07 | 1.211E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 6.008E-07 | 1.535E-04 |
|---|
| regulation of cellular component biogenesis | GO:0044087 |  | 6.821E-07 | 1.569E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 8.251E-07 | 1.725E-04 |
|---|


