Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 733
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3211 | 2.581e-03 | 6.400e-05 | 2.041e-01 | 3.371e-08 |
|---|
| IPC | 0.3616 | 3.146e-01 | 1.496e-01 | 9.285e-01 | 4.370e-02 |
|---|
| Loi | 0.4454 | 2.210e-04 | 0.000e+00 | 7.082e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5895 | 2.000e-06 | 0.000e+00 | 1.357e-01 | 0.000e+00 |
|---|
| Wang | 0.2391 | 1.535e-02 | 1.023e-01 | 3.144e-01 | 4.936e-04 |
|---|
Expression data for subnetwork 733 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
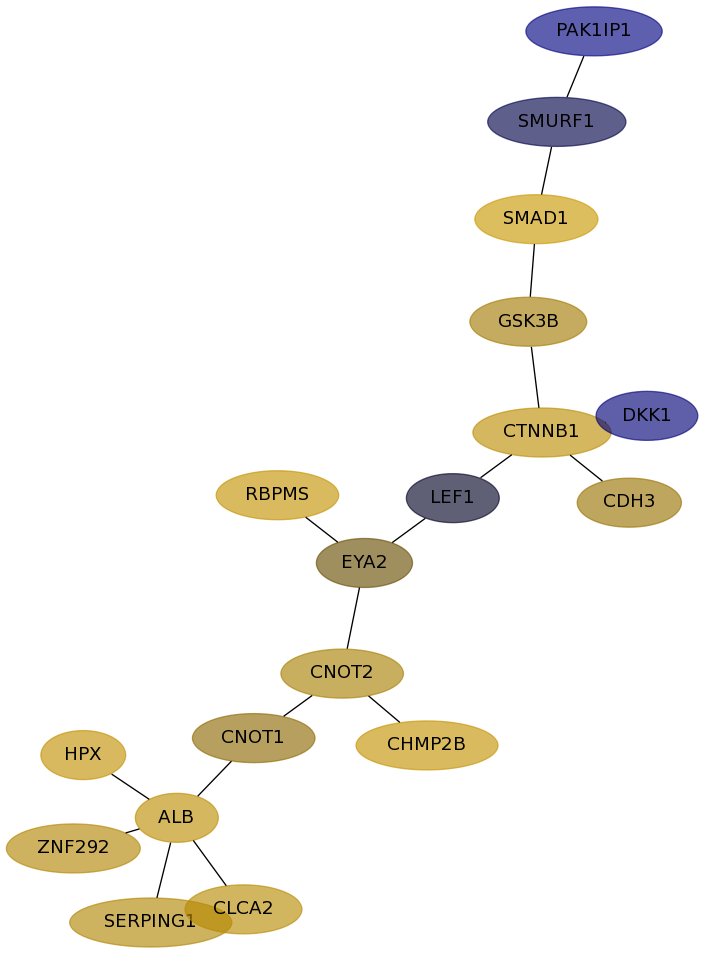
Score for each gene in subnetwork 733 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| ZNF292 |   | 1 | 652 | 1431 | 1431 | 0.165 | -0.064 | 0.164 | 0.067 | 0.055 |
|---|
| CNOT1 |   | 1 | 652 | 1431 | 1431 | 0.067 | -0.070 | 0.010 | -0.117 | 0.043 |
|---|
| CHMP2B |   | 3 | 265 | 419 | 419 | 0.250 | 0.128 | 0.134 | -0.039 | 0.067 |
|---|
| DKK1 |   | 4 | 198 | 271 | 273 | -0.041 | 0.134 | 0.091 | -0.124 | 0.129 |
|---|
| CNOT2 |   | 3 | 265 | 419 | 419 | 0.132 | 0.202 | 0.114 | 0.112 | 0.123 |
|---|
| LEF1 |   | 2 | 391 | 1325 | 1321 | -0.005 | -0.068 | 0.016 | -0.018 | 0.139 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| SERPING1 |   | 1 | 652 | 1431 | 1431 | 0.160 | -0.031 | -0.034 | -0.082 | -0.018 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| CLCA2 |   | 5 | 148 | 206 | 202 | 0.188 | -0.176 | 0.243 | -0.153 | -0.000 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| PAK1IP1 |   | 1 | 652 | 1431 | 1431 | -0.051 | 0.022 | 0.018 | 0.005 | 0.091 |
|---|
| RBPMS |   | 7 | 100 | 419 | 407 | 0.246 | 0.043 | 0.108 | 0.066 | -0.077 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| HPX |   | 1 | 652 | 1431 | 1431 | 0.236 | 0.016 | 0.070 | -0.107 | 0.005 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| EYA2 |   | 5 | 148 | 419 | 415 | 0.028 | -0.034 | 0.113 | -0.142 | -0.019 |
|---|
GO Enrichment output for subnetwork 733 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.464E-07 | 3.368E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 2.663E-07 | 3.063E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 3.085E-07 | 2.365E-04 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 4.075E-07 | 2.343E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.089E-07 | 1.881E-04 |
|---|
| gastrulation | GO:0007369 |  | 2.46E-06 | 9.431E-04 |
|---|
| mesoderm development | GO:0007498 |  | 4.27E-06 | 1.403E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.663E-05 | 7.656E-03 |
|---|
| paraxial mesoderm development | GO:0048339 |  | 3.879E-05 | 9.914E-03 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 4.216E-05 | 9.696E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.812E-05 | 0.01215146 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.231E-09 | 3.007E-06 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 5.549E-08 | 6.778E-05 |
|---|
| mesoderm formation | GO:0001707 |  | 1.312E-07 | 1.068E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 1.688E-07 | 1.031E-04 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 1.904E-07 | 9.304E-05 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.251E-06 | 5.093E-04 |
|---|
| gastrulation | GO:0007369 |  | 1.506E-06 | 5.256E-04 |
|---|
| mesoderm development | GO:0007498 |  | 2.103E-06 | 6.423E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.089E-05 | 5.671E-03 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 2.266E-05 | 5.535E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 2.854E-05 | 6.339E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.932E-10 | 2.39E-06 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.648E-08 | 7.997E-05 |
|---|
| mesoderm formation | GO:0001707 |  | 1.454E-07 | 1.166E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 1.886E-07 | 1.135E-04 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 2.135E-07 | 1.028E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.498E-06 | 6.006E-04 |
|---|
| gastrulation | GO:0007369 |  | 1.534E-06 | 5.273E-04 |
|---|
| mesoderm development | GO:0007498 |  | 2.332E-06 | 7.012E-04 |
|---|
| paraxial mesoderm development | GO:0048339 |  | 2.276E-05 | 6.083E-03 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 2.298E-05 | 5.529E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.298E-05 | 5.027E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.464E-07 | 3.368E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 2.663E-07 | 3.063E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 3.085E-07 | 2.365E-04 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 4.075E-07 | 2.343E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.089E-07 | 1.881E-04 |
|---|
| gastrulation | GO:0007369 |  | 2.46E-06 | 9.431E-04 |
|---|
| mesoderm development | GO:0007498 |  | 4.27E-06 | 1.403E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.663E-05 | 7.656E-03 |
|---|
| paraxial mesoderm development | GO:0048339 |  | 3.879E-05 | 9.914E-03 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 4.216E-05 | 9.696E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.812E-05 | 0.01215146 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.464E-07 | 3.368E-04 |
|---|
| mesoderm formation | GO:0001707 |  | 2.663E-07 | 3.063E-04 |
|---|
| formation of primary germ layer | GO:0001704 |  | 3.085E-07 | 2.365E-04 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 4.075E-07 | 2.343E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.089E-07 | 1.881E-04 |
|---|
| gastrulation | GO:0007369 |  | 2.46E-06 | 9.431E-04 |
|---|
| mesoderm development | GO:0007498 |  | 4.27E-06 | 1.403E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.663E-05 | 7.656E-03 |
|---|
| paraxial mesoderm development | GO:0048339 |  | 3.879E-05 | 9.914E-03 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 4.216E-05 | 9.696E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.812E-05 | 0.01215146 |
|---|


