Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 676
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3021 | 5.618e-03 | 2.310e-04 | 2.799e-01 | 3.632e-07 |
|---|
| IPC | 0.3952 | 2.604e-01 | 1.086e-01 | 9.166e-01 | 2.591e-02 |
|---|
| Loi | 0.4417 | 2.580e-04 | 0.000e+00 | 7.313e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6116 | 0.000e+00 | 0.000e+00 | 9.071e-02 | 0.000e+00 |
|---|
| Wang | 0.2402 | 1.472e-02 | 9.963e-02 | 3.087e-01 | 4.527e-04 |
|---|
Expression data for subnetwork 676 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
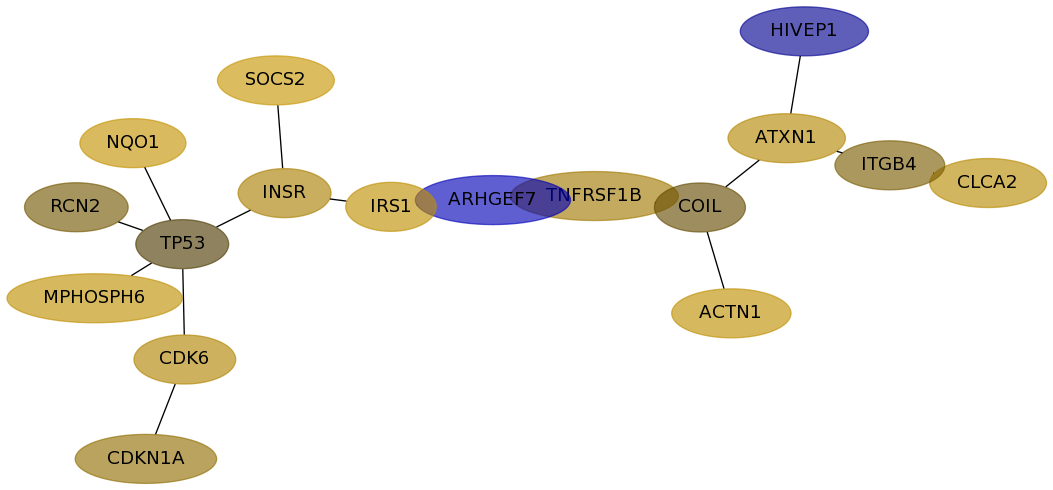
Score for each gene in subnetwork 676 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| NQO1 |   | 6 | 120 | 536 | 535 | 0.243 | -0.115 | 0.069 | -0.189 | 0.118 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| CLCA2 |   | 5 | 148 | 206 | 202 | 0.188 | -0.176 | 0.243 | -0.153 | -0.000 |
|---|
| SOCS2 |   | 4 | 198 | 859 | 837 | 0.262 | -0.130 | -0.028 | 0.051 | 0.027 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| ATXN1 |   | 6 | 120 | 364 | 355 | 0.171 | 0.026 | 0.192 | 0.175 | 0.163 |
|---|
| ACTN1 |   | 4 | 198 | 657 | 635 | 0.217 | 0.016 | 0.347 | 0.068 | 0.314 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| ITGB4 |   | 6 | 120 | 364 | 355 | 0.044 | 0.021 | 0.313 | -0.134 | -0.114 |
|---|
| COIL |   | 3 | 265 | 584 | 573 | 0.027 | 0.151 | 0.144 | 0.071 | 0.044 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| RCN2 |   | 6 | 120 | 756 | 731 | 0.037 | 0.059 | 0.051 | 0.176 | -0.098 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
| HIVEP1 |   | 6 | 120 | 419 | 410 | -0.072 | -0.199 | 0.069 | 0.000 | -0.042 |
|---|
GO Enrichment output for subnetwork 676 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.512E-08 | 1.728E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.817E-06 | 6.69E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 7.964E-06 | 6.105E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 9.209E-06 | 5.295E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.103E-05 | 5.073E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.207E-05 | 4.628E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.443E-05 | 0.01131345 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.443E-05 | 9.899E-03 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 3.443E-05 | 8.799E-03 |
|---|
| negative regulation of osteoblast differentiation | GO:0045668 |  | 3.443E-05 | 7.919E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.524E-05 | 7.368E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.457E-07 | 3.559E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 1.024E-05 | 0.01251117 |
|---|
| protein tetramerization | GO:0051262 |  | 1.024E-05 | 8.341E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.299E-05 | 7.936E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.341E-05 | 6.551E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.987E-05 | 8.092E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.793E-05 | 9.747E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.793E-05 | 8.529E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.699E-05 | 0.01003946 |
|---|
| negative regulation of neuroblast proliferation | GO:0007406 |  | 4.185E-05 | 0.01022367 |
|---|
| negative regulation of osteoblast differentiation | GO:0045668 |  | 4.185E-05 | 9.294E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.243E-07 | 2.992E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 9.029E-06 | 0.01086218 |
|---|
| protein tetramerization | GO:0051262 |  | 9.029E-06 | 7.241E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.157E-05 | 6.959E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.157E-05 | 5.568E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.62E-05 | 6.498E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.802E-05 | 9.632E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.802E-05 | 8.428E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.413E-05 | 9.124E-03 |
|---|
| male sex determination | GO:0030238 |  | 4.199E-05 | 0.0101026 |
|---|
| negative regulation of neuroblast proliferation | GO:0007406 |  | 4.199E-05 | 9.184E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.512E-08 | 1.728E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.817E-06 | 6.69E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 7.964E-06 | 6.105E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 9.209E-06 | 5.295E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.103E-05 | 5.073E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.207E-05 | 4.628E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.443E-05 | 0.01131345 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.443E-05 | 9.899E-03 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 3.443E-05 | 8.799E-03 |
|---|
| negative regulation of osteoblast differentiation | GO:0045668 |  | 3.443E-05 | 7.919E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.524E-05 | 7.368E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.512E-08 | 1.728E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.817E-06 | 6.69E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 7.964E-06 | 6.105E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 9.209E-06 | 5.295E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.103E-05 | 5.073E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.207E-05 | 4.628E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.443E-05 | 0.01131345 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.443E-05 | 9.899E-03 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 3.443E-05 | 8.799E-03 |
|---|
| negative regulation of osteoblast differentiation | GO:0045668 |  | 3.443E-05 | 7.919E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.524E-05 | 7.368E-03 |
|---|


