Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 673
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3013 | 5.795e-03 | 2.430e-04 | 2.833e-01 | 3.989e-07 |
|---|
| IPC | 0.3980 | 2.559e-01 | 1.054e-01 | 9.156e-01 | 2.470e-02 |
|---|
| Loi | 0.4415 | 2.610e-04 | 0.000e+00 | 7.329e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6120 | 0.000e+00 | 0.000e+00 | 9.012e-02 | 0.000e+00 |
|---|
| Wang | 0.2397 | 1.504e-02 | 1.010e-01 | 3.116e-01 | 4.733e-04 |
|---|
Expression data for subnetwork 673 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
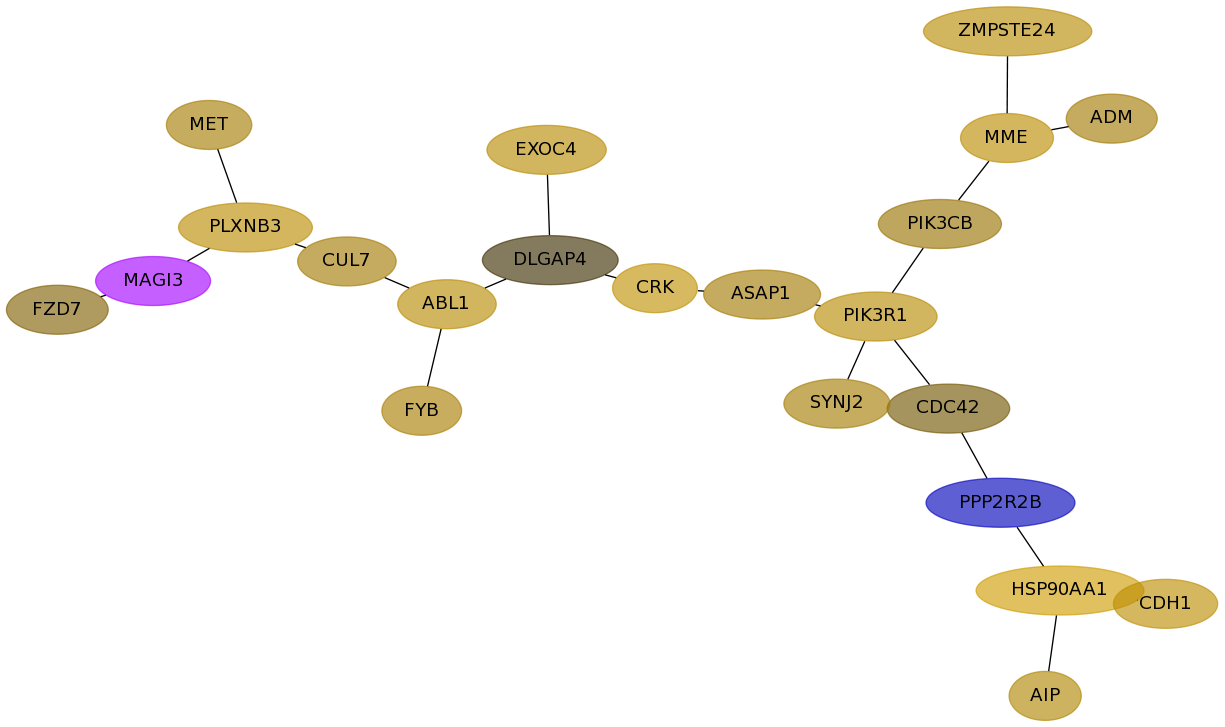
Score for each gene in subnetwork 673 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| PLXNB3 |   | 1 | 652 | 1188 | 1199 | 0.199 | -0.043 | 0.152 | -0.113 | -0.070 |
|---|
| CUL7 |   | 1 | 652 | 1188 | 1199 | 0.114 | -0.157 | 0.176 | -0.108 | 0.104 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| DLGAP4 |   | 5 | 148 | 326 | 330 | 0.010 | -0.029 | 0.180 | -0.201 | -0.043 |
|---|
| PIK3CB |   | 2 | 391 | 1188 | 1174 | 0.088 | 0.111 | 0.108 | -0.158 | 0.068 |
|---|
| AIP |   | 2 | 391 | 1188 | 1174 | 0.158 | 0.207 | -0.175 | 0.088 | -0.116 |
|---|
| MAGI3 |   | 1 | 652 | 1188 | 1199 | undef | -0.233 | 0.199 | undef | undef |
|---|
| FZD7 |   | 5 | 148 | 552 | 541 | 0.051 | -0.219 | 0.062 | 0.062 | -0.059 |
|---|
| ADM |   | 2 | 391 | 1188 | 1174 | 0.116 | 0.255 | 0.187 | 0.091 | 0.212 |
|---|
| FYB |   | 1 | 652 | 1188 | 1199 | 0.127 | -0.080 | -0.082 | -0.154 | -0.061 |
|---|
| ZMPSTE24 |   | 1 | 652 | 1188 | 1199 | 0.192 | -0.078 | 0.054 | -0.056 | 0.070 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| ASAP1 |   | 6 | 120 | 129 | 128 | 0.114 | 0.036 | 0.070 | 0.035 | -0.056 |
|---|
| EXOC4 |   | 1 | 652 | 1188 | 1199 | 0.187 | 0.008 | -0.093 | -0.124 | 0.099 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| SYNJ2 |   | 1 | 652 | 1188 | 1199 | 0.119 | -0.132 | 0.008 | 0.015 | -0.066 |
|---|
| MET |   | 5 | 148 | 536 | 536 | 0.119 | -0.219 | 0.086 | 0.183 | 0.170 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
GO Enrichment output for subnetwork 673 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 3.762E-08 | 8.653E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 5.121E-08 | 5.889E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.588E-07 | 1.218E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.588E-07 | 9.131E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.17E-07 | 1.458E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.538E-07 | 2.123E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.538E-07 | 1.82E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.324E-06 | 3.808E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.889E-06 | 4.826E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 2.592E-06 | 5.962E-04 |
|---|
| microspike assembly | GO:0030035 |  | 3.45E-06 | 7.213E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 6.911E-08 | 1.688E-04 |
|---|
| embryonic cleavage | GO:0040016 |  | 7.615E-08 | 9.302E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 8.524E-08 | 6.941E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.521E-07 | 9.289E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.521E-07 | 7.431E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.246E-07 | 1.729E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.246E-07 | 1.482E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.36E-07 | 1.942E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.246E-06 | 3.382E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.659E-06 | 4.052E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.09E-06 | 1.13E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 5.909E-08 | 1.422E-04 |
|---|
| lipid phosphorylation | GO:0046834 |  | 7.473E-08 | 8.99E-05 |
|---|
| embryonic cleavage | GO:0040016 |  | 9.565E-08 | 7.671E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.91E-07 | 1.149E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.91E-07 | 9.191E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.338E-07 | 1.338E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.338E-07 | 1.147E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.332E-07 | 1.604E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.564E-06 | 4.18E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.082E-06 | 5.009E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.266E-06 | 1.152E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 3.762E-08 | 8.653E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 5.121E-08 | 5.889E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.588E-07 | 1.218E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.588E-07 | 9.131E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.17E-07 | 1.458E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.538E-07 | 2.123E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.538E-07 | 1.82E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.324E-06 | 3.808E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.889E-06 | 4.826E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 2.592E-06 | 5.962E-04 |
|---|
| microspike assembly | GO:0030035 |  | 3.45E-06 | 7.213E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 3.762E-08 | 8.653E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 5.121E-08 | 5.889E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.588E-07 | 1.218E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.588E-07 | 9.131E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.17E-07 | 1.458E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.538E-07 | 2.123E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.538E-07 | 1.82E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.324E-06 | 3.808E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.889E-06 | 4.826E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 2.592E-06 | 5.962E-04 |
|---|
| microspike assembly | GO:0030035 |  | 3.45E-06 | 7.213E-04 |
|---|


