Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 669
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3000 | 6.091e-03 | 2.640e-04 | 2.888e-01 | 4.644e-07 |
|---|
| IPC | 0.3993 | 2.539e-01 | 1.040e-01 | 9.151e-01 | 2.416e-02 |
|---|
| Loi | 0.4408 | 2.680e-04 | 0.000e+00 | 7.372e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6143 | 0.000e+00 | 0.000e+00 | 8.609e-02 | 0.000e+00 |
|---|
| Wang | 0.2393 | 1.523e-02 | 1.017e-01 | 3.133e-01 | 4.853e-04 |
|---|
Expression data for subnetwork 669 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
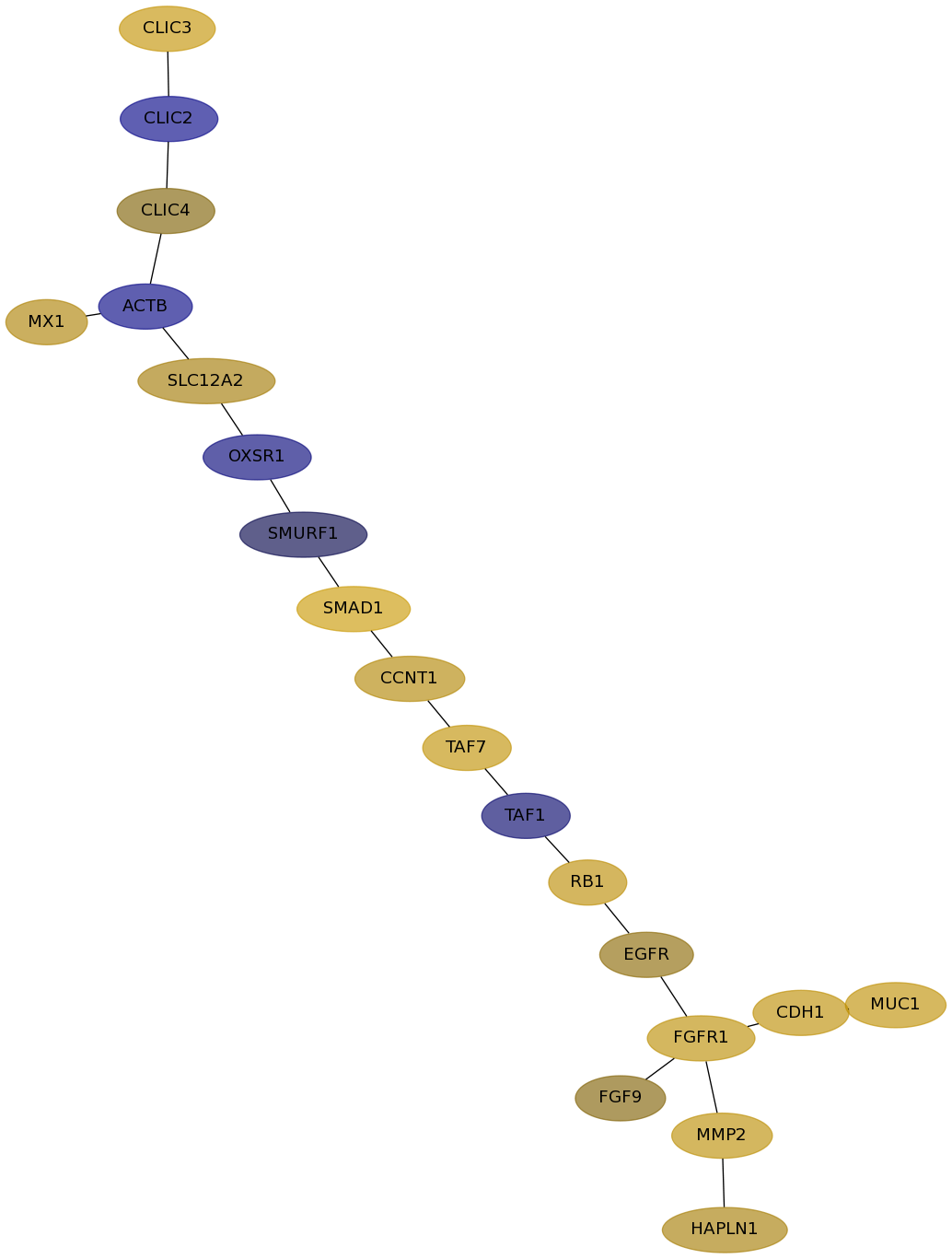
Score for each gene in subnetwork 669 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MX1 |   | 3 | 265 | 398 | 404 | 0.143 | 0.092 | -0.016 | -0.165 | -0.267 |
|---|
| FGF9 |   | 5 | 148 | 262 | 265 | 0.050 | -0.096 | 0.087 | 0.014 | 0.174 |
|---|
| TAF1 |   | 2 | 391 | 492 | 506 | -0.029 | -0.069 | 0.040 | 0.262 | -0.103 |
|---|
| CLIC2 |   | 1 | 652 | 1175 | 1182 | -0.057 | -0.123 | 0.088 | -0.081 | 0.017 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| HAPLN1 |   | 3 | 265 | 1103 | 1080 | 0.119 | 0.064 | 0.036 | 0.176 | 0.226 |
|---|
| OXSR1 |   | 2 | 391 | 994 | 983 | -0.040 | -0.109 | 0.199 | 0.223 | -0.072 |
|---|
| CLIC3 |   | 1 | 652 | 1175 | 1182 | 0.248 | -0.061 | 0.158 | -0.255 | -0.053 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CLIC4 |   | 2 | 391 | 1106 | 1100 | 0.048 | 0.141 | 0.196 | 0.282 | 0.106 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| MMP2 |   | 7 | 100 | 797 | 778 | 0.207 | -0.105 | 0.186 | -0.024 | 0.162 |
|---|
| TAF7 |   | 2 | 391 | 492 | 506 | 0.232 | 0.087 | 0.101 | -0.036 | -0.030 |
|---|
| ACTB |   | 7 | 100 | 398 | 385 | -0.052 | -0.039 | 0.283 | -0.061 | 0.003 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| SLC12A2 |   | 2 | 391 | 994 | 983 | 0.110 | 0.043 | 0.143 | -0.029 | -0.247 |
|---|
GO Enrichment output for subnetwork 669 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chloride transport | GO:0006821 |  | 8.006E-10 | 1.841E-06 |
|---|
| inorganic anion transport | GO:0015698 |  | 6.92E-09 | 7.958E-06 |
|---|
| G1 phase | GO:0051318 |  | 5.203E-06 | 3.989E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.963E-06 | 5.154E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 1.628E-05 | 7.491E-03 |
|---|
| ossification | GO:0001503 |  | 2.939E-05 | 0.01126678 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.546E-05 | 0.01165226 |
|---|
| interphase | GO:0051325 |  | 3.882E-05 | 0.01116123 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 4.582E-05 | 0.0117091 |
|---|
| regulation of fibroblast growth factor receptor signaling pathway | GO:0040036 |  | 4.582E-05 | 0.01053819 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.863E-05 | 0.01435053 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1 phase | GO:0051318 |  | 1.728E-10 | 4.221E-07 |
|---|
| chloride transport | GO:0006821 |  | 3.156E-10 | 3.855E-07 |
|---|
| inorganic anion transport | GO:0015698 |  | 3.185E-09 | 2.594E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.729E-09 | 5.331E-06 |
|---|
| G1 phase of mitotic cell cycle | GO:0000080 |  | 1.813E-08 | 8.856E-06 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.767E-08 | 1.941E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.571E-07 | 5.483E-05 |
|---|
| transcription initiation | GO:0006352 |  | 3.88E-07 | 1.185E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.22E-06 | 3.311E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.648E-06 | 4.025E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.559E-06 | 1.457E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1 phase | GO:0051318 |  | 1.173E-10 | 2.822E-07 |
|---|
| chloride transport | GO:0006821 |  | 4.182E-10 | 5.031E-07 |
|---|
| inorganic anion transport | GO:0015698 |  | 4.201E-09 | 3.369E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.192E-08 | 7.173E-06 |
|---|
| G1 phase of mitotic cell cycle | GO:0000080 |  | 1.205E-08 | 5.799E-06 |
|---|
| interphase | GO:0051325 |  | 1.439E-08 | 5.77E-06 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.235E-08 | 1.799E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.979E-07 | 5.951E-05 |
|---|
| transcription initiation | GO:0006352 |  | 5.043E-07 | 1.348E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.585E-06 | 3.815E-04 |
|---|
| RNA elongation | GO:0006354 |  | 2.153E-06 | 4.71E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chloride transport | GO:0006821 |  | 8.006E-10 | 1.841E-06 |
|---|
| inorganic anion transport | GO:0015698 |  | 6.92E-09 | 7.958E-06 |
|---|
| G1 phase | GO:0051318 |  | 5.203E-06 | 3.989E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.963E-06 | 5.154E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 1.628E-05 | 7.491E-03 |
|---|
| ossification | GO:0001503 |  | 2.939E-05 | 0.01126678 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.546E-05 | 0.01165226 |
|---|
| interphase | GO:0051325 |  | 3.882E-05 | 0.01116123 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 4.582E-05 | 0.0117091 |
|---|
| regulation of fibroblast growth factor receptor signaling pathway | GO:0040036 |  | 4.582E-05 | 0.01053819 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.863E-05 | 0.01435053 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chloride transport | GO:0006821 |  | 8.006E-10 | 1.841E-06 |
|---|
| inorganic anion transport | GO:0015698 |  | 6.92E-09 | 7.958E-06 |
|---|
| G1 phase | GO:0051318 |  | 5.203E-06 | 3.989E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.963E-06 | 5.154E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 1.628E-05 | 7.491E-03 |
|---|
| ossification | GO:0001503 |  | 2.939E-05 | 0.01126678 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.546E-05 | 0.01165226 |
|---|
| interphase | GO:0051325 |  | 3.882E-05 | 0.01116123 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 4.582E-05 | 0.0117091 |
|---|
| regulation of fibroblast growth factor receptor signaling pathway | GO:0040036 |  | 4.582E-05 | 0.01053819 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.863E-05 | 0.01435053 |
|---|


