Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 658
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2965 | 6.948e-03 | 3.290e-04 | 3.039e-01 | 6.946e-07 |
|---|
| IPC | 0.4022 | 2.494e-01 | 1.009e-01 | 9.141e-01 | 2.300e-02 |
|---|
| Loi | 0.4394 | 2.840e-04 | 0.000e+00 | 7.459e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6209 | 0.000e+00 | 0.000e+00 | 7.546e-02 | 0.000e+00 |
|---|
| Wang | 0.2391 | 1.533e-02 | 1.022e-01 | 3.143e-01 | 4.924e-04 |
|---|
Expression data for subnetwork 658 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
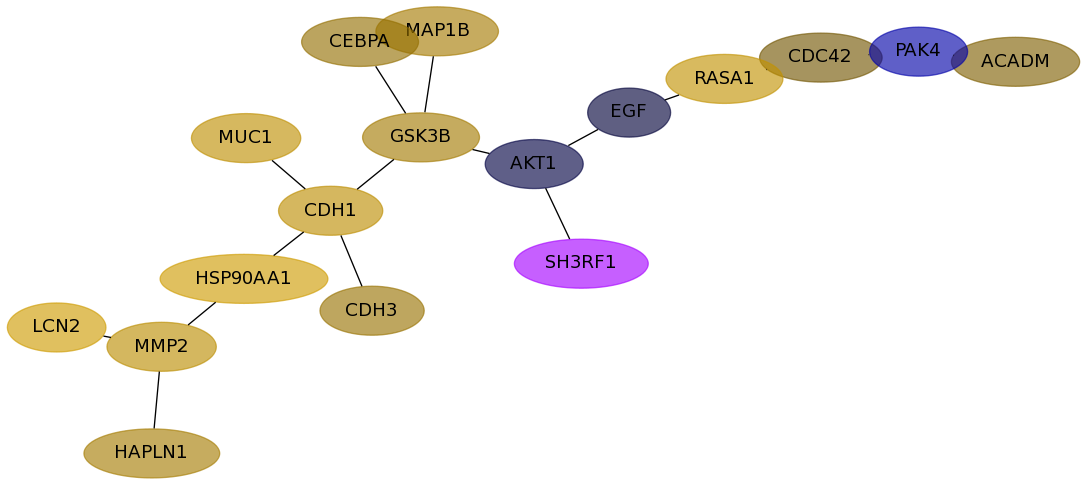
Score for each gene in subnetwork 658 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| LCN2 |   | 6 | 120 | 797 | 779 | 0.316 | 0.113 | 0.145 | -0.098 | -0.284 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| CEBPA |   | 3 | 265 | 1052 | 1036 | 0.080 | -0.058 | 0.005 | -0.100 | 0.039 |
|---|
| PAK4 |   | 1 | 652 | 1103 | 1108 | -0.130 | -0.066 | 0.084 | -0.282 | -0.072 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| HAPLN1 |   | 3 | 265 | 1103 | 1080 | 0.119 | 0.064 | 0.036 | 0.176 | 0.226 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| MMP2 |   | 7 | 100 | 797 | 778 | 0.207 | -0.105 | 0.186 | -0.024 | 0.162 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| ACADM |   | 1 | 652 | 1103 | 1108 | 0.048 | -0.012 | 0.055 | 0.047 | 0.146 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
GO Enrichment output for subnetwork 658 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.672E-08 | 8.445E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.672E-08 | 4.222E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.335E-08 | 5.624E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.282E-07 | 7.373E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.282E-07 | 5.899E-05 |
|---|
| nucleus localization | GO:0051647 |  | 3.071E-07 | 1.177E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.382E-07 | 1.44E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 6.019E-07 | 1.731E-04 |
|---|
| microspike assembly | GO:0030035 |  | 8.017E-07 | 2.049E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.575E-06 | 3.623E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.653E-06 | 3.456E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.904E-08 | 4.652E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.904E-08 | 2.326E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.324E-08 | 4.336E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.324E-08 | 3.252E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.981E-08 | 3.899E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.565E-07 | 6.374E-05 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.086E-07 | 7.28E-05 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 5.057E-07 | 1.544E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 6.425E-07 | 1.744E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 6.489E-07 | 1.585E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 7.704E-07 | 1.711E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.392E-08 | 5.754E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.392E-08 | 2.877E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.182E-08 | 3.354E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.182E-08 | 2.516E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.686E-08 | 3.218E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.966E-07 | 7.883E-05 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.619E-07 | 9.002E-05 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 5.963E-07 | 1.793E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 6.647E-07 | 1.777E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 7.712E-07 | 1.856E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.065E-07 | 1.764E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.672E-08 | 8.445E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.672E-08 | 4.222E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.335E-08 | 5.624E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.282E-07 | 7.373E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.282E-07 | 5.899E-05 |
|---|
| nucleus localization | GO:0051647 |  | 3.071E-07 | 1.177E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.382E-07 | 1.44E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 6.019E-07 | 1.731E-04 |
|---|
| microspike assembly | GO:0030035 |  | 8.017E-07 | 2.049E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.575E-06 | 3.623E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.653E-06 | 3.456E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.672E-08 | 8.445E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.672E-08 | 4.222E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.335E-08 | 5.624E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.282E-07 | 7.373E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.282E-07 | 5.899E-05 |
|---|
| nucleus localization | GO:0051647 |  | 3.071E-07 | 1.177E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.382E-07 | 1.44E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 6.019E-07 | 1.731E-04 |
|---|
| microspike assembly | GO:0030035 |  | 8.017E-07 | 2.049E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.575E-06 | 3.623E-04 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.653E-06 | 3.456E-04 |
|---|


