Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 614
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2844 | 1.084e-02 | 6.880e-04 | 3.588e-01 | 2.677e-06 |
|---|
| IPC | 0.4261 | 2.132e-01 | 7.755e-02 | 9.056e-01 | 1.498e-02 |
|---|
| Loi | 0.4388 | 2.920e-04 | 0.000e+00 | 7.503e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6307 | 0.000e+00 | 0.000e+00 | 6.160e-02 | 0.000e+00 |
|---|
| Wang | 0.2402 | 1.477e-02 | 9.985e-02 | 3.092e-01 | 4.560e-04 |
|---|
Expression data for subnetwork 614 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
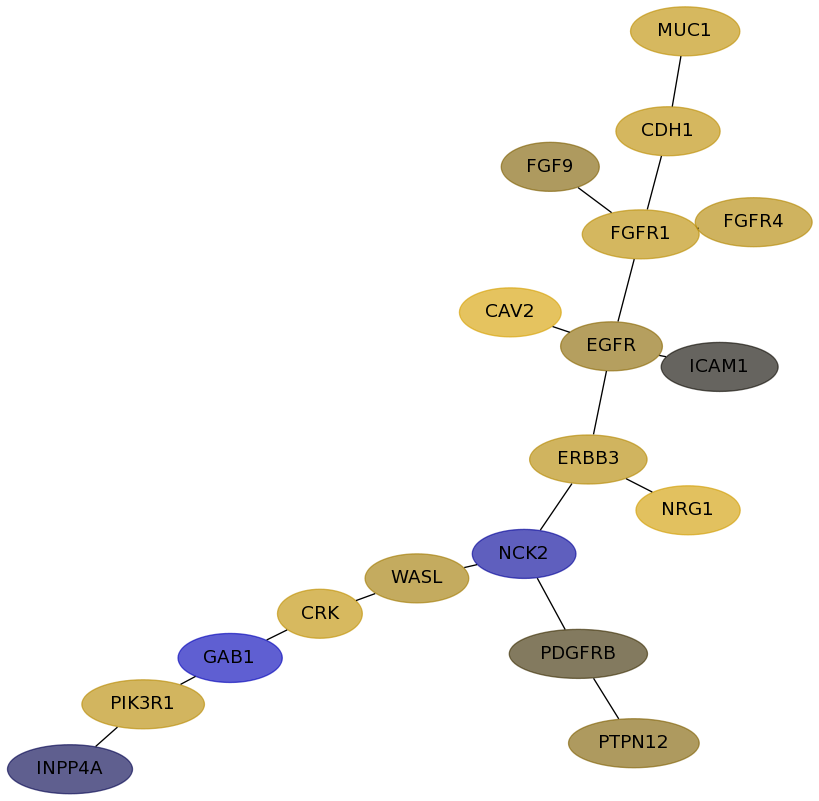
Score for each gene in subnetwork 614 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| FGF9 |   | 5 | 148 | 262 | 265 | 0.050 | -0.096 | 0.087 | 0.014 | 0.174 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PTPN12 |   | 2 | 391 | 262 | 283 | 0.048 | -0.159 | 0.153 | 0.031 | 0.042 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| ICAM1 |   | 2 | 391 | 262 | 283 | 0.001 | 0.147 | -0.054 | -0.040 | -0.132 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INPP4A |   | 1 | 652 | 262 | 310 | -0.015 | 0.091 | -0.091 | 0.027 | -0.027 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| WASL |   | 2 | 391 | 206 | 220 | 0.111 | 0.186 | -0.065 | -0.021 | -0.084 |
|---|
| NCK2 |   | 2 | 391 | 262 | 283 | -0.089 | -0.091 | 0.064 | 0.021 | -0.053 |
|---|
| NRG1 |   | 3 | 265 | 262 | 270 | 0.341 | -0.035 | 0.020 | -0.123 | -0.130 |
|---|
| ERBB3 |   | 5 | 148 | 206 | 202 | 0.175 | -0.171 | -0.007 | -0.141 | -0.098 |
|---|
| GAB1 |   | 2 | 391 | 262 | 283 | -0.192 | -0.069 | 0.084 | -0.117 | -0.212 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| PDGFRB |   | 3 | 265 | 262 | 270 | 0.009 | -0.094 | 0.224 | 0.127 | 0.189 |
|---|
GO Enrichment output for subnetwork 614 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.011E-07 | 1.383E-03 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.416E-06 | 1.629E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.787E-06 | 3.67E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.576E-06 | 5.506E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.353E-05 | 6.225E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.394E-05 | 5.344E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.604E-05 | 5.271E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 5.416E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.32E-05 | 8.484E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.32E-05 | 7.635E-03 |
|---|
| membrane docking | GO:0022406 |  | 4.771E-05 | 9.976E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.942E-09 | 4.745E-06 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.958E-07 | 2.392E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 8.108E-07 | 6.603E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.934E-06 | 1.181E-03 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 2.296E-06 | 1.122E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.53E-06 | 1.845E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 5.119E-06 | 1.786E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 6.727E-06 | 2.054E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 7.158E-06 | 1.943E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.309E-06 | 2.03E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.259E-05 | 2.797E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.581E-07 | 6.209E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 2.229E-06 | 2.681E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 5.268E-06 | 4.225E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.551E-06 | 3.339E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.794E-06 | 4.232E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 8.835E-06 | 3.543E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.185E-05 | 4.071E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.185E-05 | 3.562E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.761E-05 | 4.709E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.988E-05 | 4.784E-03 |
|---|
| membrane docking | GO:0022406 |  | 3.087E-05 | 6.752E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.011E-07 | 1.383E-03 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.416E-06 | 1.629E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.787E-06 | 3.67E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.576E-06 | 5.506E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.353E-05 | 6.225E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.394E-05 | 5.344E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.604E-05 | 5.271E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 5.416E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.32E-05 | 8.484E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.32E-05 | 7.635E-03 |
|---|
| membrane docking | GO:0022406 |  | 4.771E-05 | 9.976E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.011E-07 | 1.383E-03 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.416E-06 | 1.629E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.787E-06 | 3.67E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.576E-06 | 5.506E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.353E-05 | 6.225E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.394E-05 | 5.344E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.604E-05 | 5.271E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 5.416E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.32E-05 | 8.484E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.32E-05 | 7.635E-03 |
|---|
| membrane docking | GO:0022406 |  | 4.771E-05 | 9.976E-03 |
|---|


