Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 585
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2749 | 1.510e-02 | 1.193e-03 | 4.035e-01 | 7.267e-06 |
|---|
| IPC | 0.4409 | 1.920e-01 | 6.506e-02 | 9.005e-01 | 1.125e-02 |
|---|
| Loi | 0.4364 | 3.220e-04 | 0.000e+00 | 7.657e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6387 | 0.000e+00 | 0.000e+00 | 5.185e-02 | 0.000e+00 |
|---|
| Wang | 0.2391 | 1.536e-02 | 1.023e-01 | 3.145e-01 | 4.941e-04 |
|---|
Expression data for subnetwork 585 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
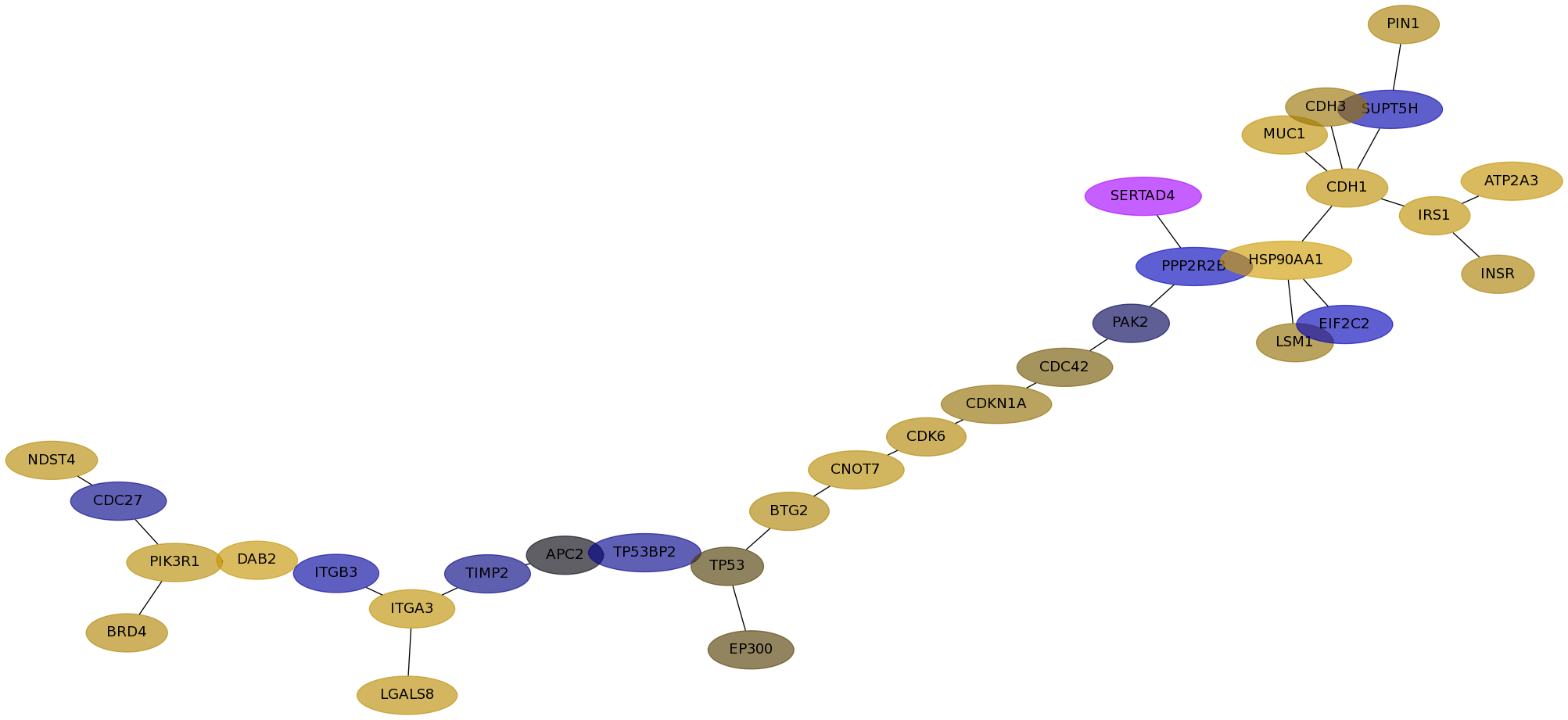
Score for each gene in subnetwork 585 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| EIF2C2 |   | 1 | 652 | 1 | 44 | -0.176 | -0.117 | -0.099 | 0.159 | 0.113 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| CNOT7 |   | 1 | 652 | 1 | 44 | 0.190 | -0.123 | -0.014 | 0.112 | -0.036 |
|---|
| APC2 |   | 1 | 652 | 1 | 44 | -0.001 | 0.029 | -0.052 | -0.078 | -0.043 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| ITGA3 |   | 3 | 265 | 1 | 24 | 0.218 | -0.135 | 0.299 | 0.037 | -0.104 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| ITGB3 |   | 3 | 265 | 1 | 24 | -0.100 | -0.015 | 0.030 | 0.086 | 0.085 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| TP53BP2 |   | 5 | 148 | 1 | 22 | -0.067 | 0.245 | -0.012 | 0.129 | 0.017 |
|---|
| LSM1 |   | 1 | 652 | 1 | 44 | 0.076 | 0.129 | -0.145 | -0.059 | 0.084 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| TIMP2 |   | 3 | 265 | 1 | 24 | -0.053 | -0.126 | 0.212 | 0.021 | 0.137 |
|---|
| LGALS8 |   | 2 | 391 | 1 | 27 | 0.197 | 0.081 | 0.002 | -0.057 | -0.071 |
|---|
| SERTAD4 |   | 7 | 100 | 1 | 20 | undef | 0.173 | 0.169 | undef | undef |
|---|
| BTG2 |   | 6 | 120 | 1 | 21 | 0.146 | -0.029 | -0.093 | -0.111 | -0.170 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CDC27 |   | 4 | 198 | 1 | 23 | -0.062 | 0.156 | -0.086 | -0.029 | 0.348 |
|---|
| NDST4 |   | 1 | 652 | 1 | 44 | 0.181 | 0.135 | -0.104 | -0.125 | 0.032 |
|---|
GO Enrichment output for subnetwork 585 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.315E-09 | 1.223E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.067E-06 | 4.907E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.541E-06 | 5.908E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 5.077E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.466E-06 | 7.089E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.689E-06 | 8.485E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.257E-06 | 1.099E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.184E-09 | 7.779E-06 |
|---|
| nuclear migration | GO:0007097 |  | 3.562E-07 | 4.351E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.562E-07 | 2.9E-04 |
|---|
| interaction with host | GO:0051701 |  | 5.56E-07 | 3.396E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 7.682E-07 | 3.753E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 8.944E-07 | 3.642E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.935E-07 | 3.467E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 9.935E-07 | 3.034E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.487E-06 | 4.037E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.487E-06 | 3.634E-04 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 2.001E-06 | 4.445E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.365E-09 | 5.691E-06 |
|---|
| nuclear migration | GO:0007097 |  | 4.79E-07 | 5.763E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.79E-07 | 3.842E-04 |
|---|
| interaction with host | GO:0051701 |  | 8.213E-07 | 4.94E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.366E-07 | 4.026E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.684E-07 | 3.883E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.134E-06 | 3.899E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.336E-06 | 4.017E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.336E-06 | 3.571E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.999E-06 | 4.81E-04 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 2.951E-06 | 6.455E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.315E-09 | 1.223E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.067E-06 | 4.907E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.541E-06 | 5.908E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 5.077E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.466E-06 | 7.089E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.689E-06 | 8.485E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.257E-06 | 1.099E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.315E-09 | 1.223E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.067E-06 | 4.907E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.541E-06 | 5.908E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 5.077E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.466E-06 | 7.089E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.689E-06 | 8.485E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.257E-06 | 1.099E-03 |
|---|


